Difference between revisions of "Pgk"
(→Biological materials) |
|||
| Line 85: | Line 85: | ||
* '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU33930] | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU33930] | ||
| − | * '''E.C. number:''' [http://www.expasy.org/enzyme/2.7.2.3 2.7.2.3] | + | * '''E.C. number:''' [http://www.expasy.org/enzyme/2.7.2.3 2.7.2.3] 2.7.2.3] |
=== Additional information=== | === Additional information=== | ||
Revision as of 11:21, 13 May 2009
- Description: phosphoglycerate kinase, glycolytic/ gluconeogenic enzyme
| Gene name | pgk |
| Synonyms | |
| Essential | yes |
| Product | phosphoglycerate kinase |
| Function | enzyme in glycolysis/ gluconeogenesis |
| MW, pI | 42,0 kDa, 4.77 |
| Gene length, protein length | 1182 bp, 394 amino acids |
| Immediate neighbours | gapA, tpi |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 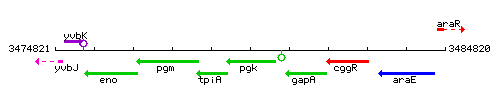
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates: 3479231 - 3480412
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + 3-phospho-D-glycerate = ADP + 1,3-bisphosphoglycerate
- Protein family: phosphoglycerate kinase family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- nucleotide binding domain (ATP) (350–353)
- 2x substrate binding domain (21–23), (59–62)
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot), Cytoplasm (Homogeneous) PubMed
Database entries
- Swiss prot entry: P40924
- KEGG entry: [3]
- E.C. number: 2.7.2.3 2.7.2.3]
Additional information
Expression and regulation
- Sigma factor: SigA
- Regulation: expression activated by glucose (2.8 fold) PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector: pGP1102 (N-terminal His-tag, in pWH844), pGP95 (N-terminal Strep-tag, in pGP172), pGP91 (N-terminal Strep-tag, for SPINE, expression in B. subtilis, in pGP380), available in Stülke lab
- lacZ fusion: pGP514 (in pAC6), a series of promoter deletion variants is also available in pAC6, available in Stülke lab
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Blencke et al. (2003) Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab Eng. 5: 133-149 PubMed
- Eymann et al. (2007) Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis. Proteomics 7: 3509-3526. PubMed
- Meile et al. (2006) Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory Proteomics 6: 2135-2146. PubMed
- Ludwig, H., Homuth, G., Schmalisch, M., Dyka, F. M., Hecker, M., and Stülke, J. (2001) Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon. Mol Microbiol 41, 409-422.PubMed
- Jannière, L., Canceill, D., Suski, C., Kanga, S., Dalmais, B., Lestini, R., Monnier, A. F., Chapuis, J., Bolotin, A., Titok, M., Le Chatelier, E., and Ehrlich, S. D. (2007) Genetic evidence for a link between glycolysis and DNA replication. PLoS ONE 2, e447. PubMed
- Macek et al. (2007) The serine/ threonine/ tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6: 697-707 PubMed