Difference between revisions of "XylA"
| Line 95: | Line 95: | ||
* '''Sigma factor:''' [[SigA]] | * '''Sigma factor:''' [[SigA]] | ||
| − | * '''Regulation:''' repressed by glucose ([[CcpA]]) | + | * '''Regulation:''' repressed by glucose ([[CcpA]]) , carbon catabolite repression ([[CcpA]]), induction by xylose ([[XylR]]) |
* '''Regulatory mechanism:''' [[CcpA]]: transcription repression, [[XylR]]: transcription repression | * '''Regulatory mechanism:''' [[CcpA]]: transcription repression, [[XylR]]: transcription repression | ||
| Line 123: | Line 123: | ||
=References= | =References= | ||
| − | # Blencke et al. (2003) Transcriptional profiling of gene expression in response to glucose in ''Bacillus subtilis'': regulation of the central metabolic pathways. ''Metab Eng.'' '''5:''' 133-149 | + | # Blencke et al. (2003) Transcriptional profiling of gene expression in response to glucose in ''Bacillus subtilis'': regulation of the central metabolic pathways. ''Metab Eng.'' '''5:''' 133-149 |
# Dahl, M. K., J. Degenkolb, and W. Hillen. 1994. Transcription of the xyl operon is controlled in Bacillus subtilis by tandem overlapping operators spaced by four base-pairs. J. Mol. Biol. 243:413-424. [http://www.ncbi.nlm.nih.gov/sites/entrez/7966270 PubMed] | # Dahl, M. K., J. Degenkolb, and W. Hillen. 1994. Transcription of the xyl operon is controlled in Bacillus subtilis by tandem overlapping operators spaced by four base-pairs. J. Mol. Biol. 243:413-424. [http://www.ncbi.nlm.nih.gov/sites/entrez/7966270 PubMed] | ||
# Jacob S, Allmansberger R, Gärtner D, Hillen W (1991) Catabolite repression of the operon for xylose utilization from Bacillus subtilis W23 is mediated at the level of transcription and depends on a cis site in the xylA reading frame. Mol Gen Genet 229: 189-196. [http://www.ncbi.nlm.nih.gov/sites/entrez/1921970 PubMed] | # Jacob S, Allmansberger R, Gärtner D, Hillen W (1991) Catabolite repression of the operon for xylose utilization from Bacillus subtilis W23 is mediated at the level of transcription and depends on a cis site in the xylA reading frame. Mol Gen Genet 229: 189-196. [http://www.ncbi.nlm.nih.gov/sites/entrez/1921970 PubMed] | ||
Revision as of 23:28, 2 April 2009
- Description: xylose isomerase
| Gene name | xylA |
| Synonyms | |
| Essential | no |
| Product | xylose isomerase |
| Function | utilization of xylan and xylose |
| MW, pI | 50 kDa, 5.635 |
| Gene length, protein length | 1335 bp, 445 aa |
| Immediate neighbours | xylR, xylB |
| Gene sequence (+200bp) | Protein sequence |
| Caution: The sequence for this gene in SubtiList contains errors | |
Genetic context 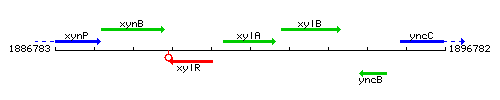
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry: [3]
- E.C. number: 5.3.1.5
Additional information
Expression and regulation
- Sigma factor: SigA
- Regulation: repressed by glucose (CcpA) , carbon catabolite repression (CcpA), induction by xylose (XylR)
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Wolfgang Hillen, Erlangen University, Germany Homepage
Your additional remarks
References
- Blencke et al. (2003) Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways. Metab Eng. 5: 133-149
- Dahl, M. K., J. Degenkolb, and W. Hillen. 1994. Transcription of the xyl operon is controlled in Bacillus subtilis by tandem overlapping operators spaced by four base-pairs. J. Mol. Biol. 243:413-424. PubMed
- Jacob S, Allmansberger R, Gärtner D, Hillen W (1991) Catabolite repression of the operon for xylose utilization from Bacillus subtilis W23 is mediated at the level of transcription and depends on a cis site in the xylA reading frame. Mol Gen Genet 229: 189-196. PubMed
- Kraus A, Hueck C, Gärtner D, Hillen W (1994) Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J Bacteriol 176: 1738-1745. PubMed
- Gärtner D, Geißendörfer M, Hillen W. 1988. Expression of the Bacillus subtilis xyl operon is repressed at the level of transcription and is induced by xylose. J. Bacteriol. 170:3102-3109. PubMed
- Gärtner D, Degenkolb J, Ripperger JAE, Allmansberger R, Hillen W. 1992. Regulation of the Bacillus subtilis W23 xylose utilization operon: interaction of Xyl repressor with xyl operator and the inducer xylose. Mol. Gen. Genet. 232:415-422 PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed