Difference between revisions of "Aag"
| Line 91: | Line 91: | ||
* '''Operon:''' | * '''Operon:''' | ||
| − | * '''Sigma factor:''' | + | * '''Sigma factor:''' ''[[SigB]]'' [http://www.ncbi.nlm.nih.gov/pubmed/15805528 PubMed] |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 119: | Line 119: | ||
=References= | =References= | ||
| + | # Höper et al. (2005) Comprehensive Characterization of the Contribution of Individual ''[[SigB]]''-Dependent General Stress Genes to Stress Resistance of ''Bacillus subtilis''. J. Bact. '''187:''' 2810-2826 [http://www.ncbi.nlm.nih.gov/pubmed/15805528 PubMed] | ||
# Aamodt et al. (2004) The ''Bacillus subtilis'' counterpart of the mammalian 3-methyladenine DNA glycosylase has hypoxanthine and 1,N6-ethenoadenine as preferred substrates. ''J. Biol. Chem.'' '''279:''' 13601-13606-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/14729667 PubMed] | # Aamodt et al. (2004) The ''Bacillus subtilis'' counterpart of the mammalian 3-methyladenine DNA glycosylase has hypoxanthine and 1,N6-ethenoadenine as preferred substrates. ''J. Biol. Chem.'' '''279:''' 13601-13606-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/14729667 PubMed] | ||
Revision as of 16:48, 27 March 2009
- Description: general stress protein, similar to DNA-3-methyladenine glycosidase II
| Gene name | aag |
| Synonyms | yxlJ |
| Essential | no |
| Product | unknown |
| Function | survival of salt and ethanol stresses |
| MW, pI | 21 kDa, 6.117 |
| Gene length, protein length | 588 bp, 196 aa |
| Immediate neighbours | yxzF, katX |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 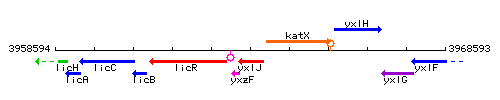
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Coordinates:
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: removes hypoxanthine, 3-alkylated purines, and 1,N6-ethenoadenine from DNA; hypoxanthine and 1,N6-ethenoadenine are the preferred substrates PubMed
- Protein family: AAG family of 3-methyladenine DNA glycosylases
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry:
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
- Höper et al. (2005) Comprehensive Characterization of the Contribution of Individual SigB-Dependent General Stress Genes to Stress Resistance of Bacillus subtilis. J. Bact. 187: 2810-2826 PubMed
- Aamodt et al. (2004) The Bacillus subtilis counterpart of the mammalian 3-methyladenine DNA glycosylase has hypoxanthine and 1,N6-ethenoadenine as preferred substrates. J. Biol. Chem. 279: 13601-13606-page. PubMed