Difference between revisions of "PckA"
(→Database entries) |
|||
| Line 83: | Line 83: | ||
* '''Swiss prot entry:''' [http://www.expasy.ch/cgi-bin/sprot-search-ac?P54418] | * '''Swiss prot entry:''' [http://www.expasy.ch/cgi-bin/sprot-search-ac?P54418] | ||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU30560] |
* '''E.C. number:''' [http://www.expasy.ch/cgi-bin/get-enzyme-entry?4.1.1.49] | * '''E.C. number:''' [http://www.expasy.ch/cgi-bin/get-enzyme-entry?4.1.1.49] | ||
Revision as of 10:31, 20 March 2009
- Description: phosphoenolpyruvate carboxykinase
| Gene name | pckA |
| Synonyms | ppc |
| Essential | no |
| Product | phosphoenolpyruvate carboxykinase |
| Function | synthesis of phosphoenolpyruvate |
| MW, pI | 58,1 kDa, 5.12 |
| Gene length, protein length | 1581 bp, 527 amino acids |
| Immediate neighbours | metK, ytmB |
| Gene sequence (+200bp) | Protein sequence |
| Caution: The sequence for this gene in SubtiList contains errors | |
Genetic context 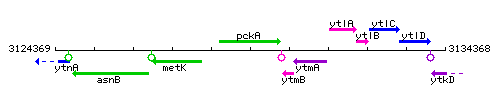
| |
Contents
The gene
Basic information
- Coordinates: 3128579 - 3130159
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + oxaloacetate = ADP + phosphoenolpyruvate + CO(2)
- Protein family: phosphoenolpyruvate carboxykinase [ATP] family
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Nucleotide binding Domain (233–240)
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm
Database entries
- Structure: 2PXZ (E.coli)
- Swiss prot entry: [3]
- KEGG entry: [4]
- E.C. number: [5]
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism: transcription repression
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- Antibody:
Labs working on this gene/protein
Stephane Aymerich, Microbiology and Molecular Genetics, INRA Paris-Grignon, France