|
|
| Line 1: |
Line 1: |
| − | * '''Description:''' transcriptional regulator of transition state genes <br/><br/> | + | * '''Description:''' write here <br/><br/> |
| | | | |
| | {| align="right" border="1" cellpadding="2" | | {| align="right" border="1" cellpadding="2" |
| | |- | | |- |
| | |style="background:#ABCDEF;" align="center"|'''Gene name''' | | |style="background:#ABCDEF;" align="center"|'''Gene name''' |
| − | |''abrB'' | + | |''aadK'' |
| | |- | | |- |
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''cpsX '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' '' |
| | |- | | |- |
| | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | | |style="background:#ABCDEF;" align="center"| '''Essential''' || no |
| | |- | | |- |
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || transcriptional regulator | + | |style="background:#ABCDEF;" align="center"| '''Product''' || aminoglycoside 6-adenylyltransferase (EC 2.7.7.-) |
| | |- | | |- |
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of gene expression during the transition from growth to stationary phase | + | |style="background:#ABCDEF;" align="center"|'''Function''' || |
| | |- | | |- |
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 10 kDa, 6.57 | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 33 kDa, 4.755 |
| | |- | | |- |
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 288 bp, 96 aa | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 852 bp, 284 aa |
| | |- | | |- |
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yabC]]'', ''[[metS]]'' | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yrdA]]'', ''[[yrpB]]'' |
| | |- | | |- |
| − | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/abrB_nucleotide.txt Gene sequence (+200bp) ]''' | + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/aadK_nucleotide.txt Gene sequence (+200bp) ]''' |
| − | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/abrB_protein.txt Protein sequence]''' | + | |style="background:#FAF8CC;" align="center"|'''[http://subtiwiki.uni-goettingen.de/aadK_protein.txt Protein sequence]''' |
| | |- | | |- |
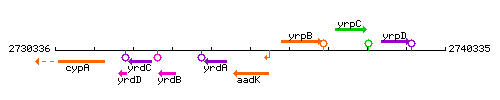
| − | |colspan="2" | '''Genetic context''' <br/> [[Image:abrB_context.gif]] | + | |colspan="2" | '''Genetic context''' <br/> [[Image:aadK_context.gif]] |
| | |- | | |- |
| | |} | | |} |
| Line 38: |
Line 38: |
| | | | |
| | ===Phenotypes of a mutant === | | ===Phenotypes of a mutant === |
| − |
| |
| − | No swarming motility on B medium. [http://www.ncbi.nlm.nih.gov/sites/entrez/19202088 PubMed]
| |
| | | | |
| | === Database entries === | | === Database entries === |
| | | | |
| − | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/abrB.html] | + | * '''DBTBS entry:''' no entry |
| | | | |
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10100] | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG11037] |
| | | | |
| | === Additional information=== | | === Additional information=== |
| Line 58: |
Line 56: |
| | * '''Protein family:''' | | * '''Protein family:''' |
| | | | |
| − | * '''Paralogous protein(s):''' [[Abh]], [[SpoVT]] (only N-terminal domain) | + | * '''Paralogous protein(s):''' |
| − | | |
| − | === Genes/ operons controlled by AbrB ===
| |
| − | | |
| − | * '''Activated by AbrB:''' ''[[citB]]'', ''[[comK]], [[hpr]]'', ''[[rbsR]]-[[rbsK]]-[[rbsD]]-[[rbsA]]-[[rbsC]]-[[rbsB]]''
| |
| − | | |
| − | * ''' Repressed by AbrB:''' ''[[abrB]], [[aprE]], [[ftsA]]-[[ftsZ]], [[kinC]], [[motA]], [[nprE]], [[pbpE]], [[spo0H]], [[spoVG]], [[spo0E]], [[tycA]], [[sbo]]-[[alb]], [[yqxM]]-[[sipW]]-[[tasA]]''
| |
| | | | |
| | === Extended information on the protein === | | === Extended information on the protein === |
| Line 76: |
Line 68: |
| | * '''Cofactor(s):''' | | * '''Cofactor(s):''' |
| | | | |
| − | * '''Effectors of protein activity:''' interaction with [[AbbA]] results in inactivation of AbrB [http://www.ncbi.nlm.nih.gov/sites/entrez/18840696 PubMed] | + | * '''Effectors of protein activity:''' |
| | | | |
| − | * '''Interactions:''' [[AbrB]]-[[AbbA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/18840696 PubMed] | + | * '''Interactions:''' |
| | | | |
| | * '''Localization:''' | | * '''Localization:''' |
| Line 84: |
Line 76: |
| | === Database entries === | | === Database entries === |
| | | | |
| − | * '''Structure:''' 1Z0R (N-terminal DNA recognition domain) [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=32611 NCBI] [http://www.ncbi.nlm.nih.gov/sites/entrez/16223496 PubMed] | + | * '''Structure:''' |
| | | | |
| | * '''Swiss prot entry:''' | | * '''Swiss prot entry:''' |
| | | | |
| − | * '''KEGG entry:''' | + | * '''KEGG entry:''' http://www.genome.jp/dbget-bin/www_bget?bsu+BSU26790 |
| | + | |
| | + | |
| | + | * '''E.C. number:''' |
| | | | |
| | === Additional information=== | | === Additional information=== |
| − |
| |
| | | | |
| | =Expression and regulation= | | =Expression and regulation= |
| | | | |
| − | * '''Operon:''' ''abrB'' [http://www.ncbi.nlm.nih.gov/sites/entrez/3145384 PubMed] | + | * '''Operon:''' |
| | | | |
| − | * '''Sigma factor:''' [[SigA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/3145384 PubMed] | + | * '''Sigma factor:''' |
| | | | |
| − | * '''Regulation:''' expressed at the onset of stationary phase [http://www.ncbi.nlm.nih.gov/sites/entrez/3145384 PubMed] | + | * '''Regulation:''' |
| | | | |
| − | * '''Regulatory mechanism:''' repressed by [[Spo0A]]-P [http://www.ncbi.nlm.nih.gov/sites/entrez/3145384 PubMed] | + | * '''Regulatory mechanism:''' |
| | | | |
| − | * '''Additional information:''' | + | * '''Additional information:''' |
| | | | |
| | =Biological materials = | | =Biological materials = |
| | | | |
| − | * '''Mutant:''' TT731 (aphA3) | + | * '''Mutant:''' |
| | | | |
| | * '''Expression vector:''' | | * '''Expression vector:''' |
| Line 120: |
Line 114: |
| | | | |
| | =Labs working on this gene/protein= | | =Labs working on this gene/protein= |
| − |
| |
| − | [[Richard Losick]], Harvard Univ., Cambridge, USA [http://www.mcb.harvard.edu/Losick/ homepage]
| |
| − |
| |
| − | [[Mark Strauch]], Baltimore, USA [http://lifesciences.umaryland.edu/Pages/faculty_profile.aspx?ID=212 homepage]
| |
| | | | |
| | =Your additional remarks= | | =Your additional remarks= |
| Line 129: |
Line 119: |
| | =References= | | =References= |
| | | | |
| − | # Banse et al. (2008) Parallel pathways of repression and antirepression governing the transition to stationary phase in ''Bacillus subtilis''.''Proc. Natl. Acad. Sci. USA'' '''105:''' 15547-15552. [http://www.ncbi.nlm.nih.gov/sites/entrez/18840696 PubMed]
| |
| − | # Perego et al. (1988) Structure of the gene for the transition state regulator, ''abrB'': regulator synthesis is controlled by the ''spo0A'' sporulation gene in ''Bacillus subtilis''. ''Mol. Microbiol.'' '''2:''' 689-699. [http://www.ncbi.nlm.nih.gov/sites/entrez/3145384 PubMed]
| |
| − | # Xu, K. and M.A. Strauch. (1996) In vitro selection of optimal AbrB-binding sites: comparison to known in vivo sites indicates flexibility in AbrB-binding and recognition of three-dimensional DNA structures. Molec. Microbiol. 19: 145-158 [http://www.ncbi.nlm.nih.gov/sites/entrez/8821944 PubMed]
| |
| − | # Xu, K., D. Clark and M.A. Strauch. (1996) Analysis of abrB mutations, mutant proteins, and why abrB does not utilize a perfect consensus in the –35 region of its sigmaA promoter.J. Biol. Chem. 271:2621-2626 [http://www.ncbi.nlm.nih.gov/sites/entrez/8576231 PubMed]
| |
| − | # Vaughn, J.L., Feher V., Naylor, S., Strauch, M.A. and J. Cavanagh. (2000) Novel DNA binding domain and genetic regulation model of Bacillus subtilis transition state regulator AbrB. Nature Structural Biology 7:1139-1146; [http://www.ncbi.nlm.nih.gov/sites/entrez/11101897 PubMed], Corrigendum appears in Nature Stuctural & Molecular Biology (2005) 12:380
| |
| − | # Xu, K. and M.A. Strauch. (2001) DNA-binding activity of amino-terminal domains of the Bacillus subtilis AbrB protein. J. Bacteriol. 183:4094-4098 [http://www.ncbi.nlm.nih.gov/sites/entrez/11395475 PubMed]
| |
| − | # Phillips, Z. E.V. and M.A. Strauch. (2001) Role of Cys54 in AbrB multimerization and DNA-binding activity. FEMS Microbiol. Letters. 203:207-210 [http://www.ncbi.nlm.nih.gov/sites/entrez/11583849 PubMed]
| |
| − | # Phillips, Z.E.V. and M.A. Strauch. (2002) Bacillus subtilis sporulation and stationary phase gene expression. Cellular and Molecular Life Sciences 59:392-402 [http://www.ncbi.nlm.nih.gov/sites/entrez/11964117 PubMed]
| |
| − | # Shafikhani, S.H., Mandic-Mulec, I., Strauch, M.A., Smith, I. and T. Leighton. (2002) Postexponential regulation of sin operon expression in Bacillus subtilis. J. Bacteriol. 184:564-571 [http://www.ncbi.nlm.nih.gov/sites/entrez/11751836 PubMed]
| |
| − | # Benson, L. M., Vaughn, J. L., Strauch, M. A., Bobay, B. G., Thompson, R., Naylor, S. and J. Cavanagh (2002). Macromolecular assembly of the transition state regulator AbrB in its unbound and complexed states probed by microelectrospray ionization mass spectrometry. Analytical Biochemistry 306:222-227 [http://www.ncbi.nlm.nih.gov/sites/entrez/12123659 PubMed]
| |
| − | # Qian, Q., Lee, C.Y., Helmann, J. and M.A. Strauch. (2002) AbrB regulation of the sigmaW regulon of Bacillus subtilis. FEMS Microbiol. Letters 211:219-223. [http://www.ncbi.nlm.nih.gov/sites/entrez/12076816 PubMed]
| |
| − | # Kim, H. J., S. I. Kim, M. Ratnayake-Lecamwasam, K. Tachikawa, A. L. Sonenshein, and M. Strauch. (2003) Complex regulation of the Bacillus subtilis aconitase gene. J. Bacteriol. 185:1672-1680. [http://www.ncbi.nlm.nih.gov/sites/entrez/12591885 PubMed]
| |
| − | # Bobay, B.G., Benson, L., Naylor, S., Feeney, B., Clark, A.C., Goshe, M.B., Strauch, M.A., Thompson, R., and J.Cavanagh. (2004) Evaluation of the DNA binding tendencies of the transition state regulator AbrB. Biochemistry. 43:16106-16118. [http://www.ncbi.nlm.nih.gov/sites/entrez/15610005 PubMed]
| |
| − | # Bobay et al.(2005) Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins. FEBS Lett. 579:5669-5674. [http://www.ncbi.nlm.nih.gov/sites/entrez/16223496 PubMed]
| |
| − | # Yao, F and M.A. Strauch (2005) Independent and Interchangeable Multimerization Domains of the AbrB, Abh and SpoVT Global Regulatory Proteins. J. Bacteriol. 187:6354-6362 [http://www.ncbi.nlm.nih.gov/sites/entrez/16159768 PubMed]
| |
| − | # Bobay, B.G., Mueller, G.A., Thompson, R.J., Venters, R.A., Murzin, A.G., Strauch, M.A. & J. Cavanagh (2006) NMR structure of AbhN and comparison with AbrBN: First Insights into the DNA-binding Promiscuity and Specificity of AbrB-like Transition-state Regulator Proteins. J. Biol. Chem. 281:21399-21409 [http://www.ncbi.nlm.nih.gov/sites/entrez/16702211 PubMed]
| |
| − | # Jordan S. Rietkötter E. Strauch MA. Kalamorz F. Butcher BG. Helmann JD. Mascher T. (2007) LiaRS-dependent gene expression is embedded in transition state regulation in Bacillus subtilis. Microbiology. 153: 2530-2540. [http://www.ncbi.nlm.nih.gov/sites/entrez/17660417 PubMed]
| |
| − | # Strauch MA. Bobay BG. Cavanagh J. Yao F. Wilson A. Le Breton Y. (2007) Abh and AbrB control of Bacillus subtilis antimicrobial gene expression. J. of Bacteriol. 189:7720-7732. [http://www.ncbi.nlm.nih.gov/sites/entrez/17720793 PubMed]
| |
| − | # Hamze et al. (2009) Identification of genes required for different stages of dendritic swarming in ''Bacillus subtilis'', with a novel role for ''phrC''. ''Microbiology'' '''155:''' 398-412. [http://www.ncbi.nlm.nih.gov/sites/entrez/19202088 PubMed]
| |
| − | # Strauch MA. (1995) AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon. ''J Bacteriol.'' '''Dec;177(23):'''6727-31. [http://www.ncbi.nlm.nih.gov/sites/entrez/7592460 PubMed]
| |
| | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] |