Difference between revisions of "YumC"
| Line 118: | Line 118: | ||
** number of protein molecules per cell (minimal medium with glucose and ammonium): 5753 {{PubMed|24696501}} | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 5753 {{PubMed|24696501}} | ||
** number of protein molecules per cell (complex medium with amino acids, without glucose): 9972 {{PubMed|24696501}} | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 9972 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 5216 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 2849 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 3293 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
Revision as of 14:21, 17 April 2014
- Description: ferredoxin-NAD(P)+ oxidoreductase
| Gene name | yumC |
| Synonyms | |
| Essential | yes PubMed |
| Product | ferredoxin-NAD(P)+ oxidoreductase |
| Function | redox reactions that involve ferredoxin |
| Gene expression levels in SubtiExpress: yumC | |
| MW, pI | 36 kDa, 5.573 |
| Gene length, protein length | 996 bp, 332 aa |
| Immediate neighbours | yumB, yuzG |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 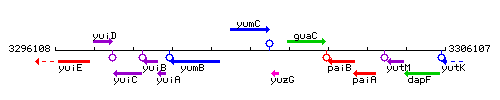
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
electron transport/ other, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU32110
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU32110
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2 reduced ferredoxin + NADP+ + H+ = 2 oxidized ferredoxin + NADPH (according to Swiss-Prot)
- Protein family: ferredoxin--NADP reductase type 2 family (according to Swiss-Prot)
- Paralogous protein(s): YcgT
Extended information on the protein
- Kinetic information:
- Modification: active site Cys85 is S-bacillithiolated by NaOCl stress in B. subtilis and other Bacillus speciesPubMed
- Effectors of protein activity:
- Interactions:
- dimeric protein PubMed
Database entries
- BsubCyc: BSU32110
- UniProt: O05268
- KEGG entry: [2]
- E.C. number: 1.18.1.2
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 5753 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 9972 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 5216 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 2849 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 3293 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Daisuke Seo, Tomoya Asano, Hirofumi Komori, Takeshi Sakurai
Role of the C-terminal extension stacked on the re-face of the isoalloxazine ring moiety of the flavin adenine dinucleotide prosthetic group in ferredoxin-NADP(+) oxidoreductase from Bacillus subtilis.
Plant Physiol Biochem: 2014, 81;143-8
[PubMed:24529496]
[WorldCat.org]
[DOI]
(I p)
Bui Khanh Chi, Alexandra A Roberts, Tran Thi Thanh Huyen, Katrin Bäsell, Dörte Becher, Dirk Albrecht, Chris J Hamilton, Haike Antelmann
S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria.
Antioxid Redox Signal: 2013, 18(11);1273-95
[PubMed:22938038]
[WorldCat.org]
[DOI]
(I p)
Hirofumi Komori, Daisuke Seo, Takeshi Sakurai, Yoshiki Higuchi
Crystal structure analysis of Bacillus subtilis ferredoxin-NADP(+) oxidoreductase and the structural basis for its substrate selectivity.
Protein Sci: 2010, 19(12);2279-90
[PubMed:20878669]
[WorldCat.org]
[DOI]
(I p)
Hirofumi Komori, Daisuke Seo, Takeshi Sakurai, Yoshiki Higuchi
Crystallization and preliminary X-ray studies of ferredoxin-NADP+ oxidoreductase encoded by Bacillus subtilis yumC.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2010, 66(Pt 3);301-3
[PubMed:20208166]
[WorldCat.org]
[DOI]
(I p)
Daisuke Seo, Seisuke Okabe, Mitsuhiro Yanase, Kunishige Kataoka, Takeshi Sakurai
Studies of interaction of homo-dimeric ferredoxin-NAD(P)+ oxidoreductases of Bacillus subtilis and Rhodopseudomonas palustris, that are closely related to thioredoxin reductases in amino acid sequence, with ferredoxins and pyridine nucleotide coenzymes.
Biochim Biophys Acta: 2009, 1794(4);594-601
[PubMed:19162251]
[WorldCat.org]
[DOI]
(P p)
Daisuke Seo, Kei Kamino, Kazuhito Inoue, Hidehiro Sakurai
Purification and characterization of ferredoxin-NADP+ reductase encoded by Bacillus subtilis yumC.
Arch Microbiol: 2004, 182(1);80-9
[PubMed:15252706]
[WorldCat.org]
[DOI]
(P p)