Difference between revisions of "MinD"
| Line 129: | Line 129: | ||
** number of protein molecules per cell (minimal medium with glucose and ammonium): 716 {{PubMed|24696501}} | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 716 {{PubMed|24696501}} | ||
** number of protein molecules per cell (complex medium with amino acids, without glucose): 813 {{PubMed|24696501}} | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 813 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 3544 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 1500 {{PubMed|21395229}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 6195 {{PubMed|21395229}} | ||
=Biological materials = | =Biological materials = | ||
| − | |||
* '''Mutant:''' | * '''Mutant:''' | ||
Revision as of 14:10, 17 April 2014
- Description: cell-division inhibitor (septum placement), part of the Min system (with DivIVA, MinC, MinJ), Noc and the Min system ensure the efficient utilization of the division site at midcell in by ensuring Z ring placement
| Gene name | minD |
| Synonyms | divIVB1 |
| Essential | no |
| Product | cell-division inhibitor |
| Function | septum placement |
| Gene expression levels in SubtiExpress: minD | |
| Interactions involving this protein in SubtInteract: MinD | |
| MW, pI | 29 kDa, 4.984 |
| Gene length, protein length | 804 bp, 268 aa |
| Immediate neighbours | spoIVFA, minC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 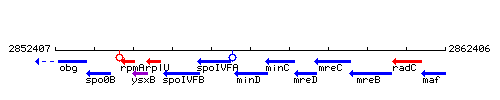
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell division, cell envelope stress proteins (controlled by SigM, V, W, X, Y), membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU27990
Phenotypes of a mutant
Database entries
- BsubCyc: BSU27990
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: The Min system prevents minicell formation adjacent to recently completed division sites by promoting the disassembly of the cytokinetic ring, thereby ensuring that cell division occurs only once per cell cycle PubMed
- Protein family: MinD subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- BsubCyc: BSU27990
- Structure:
- UniProt: Q01464
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium): 716 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 813 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 3544 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 1500 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 6195 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Imrich Barák
Open questions about the function and evolution of bacterial Min systems.
Front Microbiol: 2013, 4;378
[PubMed:24367361]
[WorldCat.org]
[DOI]
(P e)
Marc Bramkamp, Suey van Baarle
Division site selection in rod-shaped bacteria.
Curr Opin Microbiol: 2009, 12(6);683-8
[PubMed:19884039]
[WorldCat.org]
[DOI]
(I p)
Original Publications
Katarina Surdova, Pamela Gamba, Dennis Claessen, Tjalling Siersma, Martijs J Jonker, Jeff Errington, Leendert W Hamoen
The conserved DNA-binding protein WhiA is involved in cell division in Bacillus subtilis.
J Bacteriol: 2013, 195(24);5450-60
[PubMed:24097947]
[WorldCat.org]
[DOI]
(I p)
Christopher D A Rodrigues, Elizabeth J Harry
The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization.
PLoS Genet: 2012, 8(3);e1002561
[PubMed:22457634]
[WorldCat.org]
[DOI]
(I p)
Veronica Guariglia-Oropeza, John D Helmann
Bacillus subtilis σ(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids.
J Bacteriol: 2011, 193(22);6223-32
[PubMed:21926231]
[WorldCat.org]
[DOI]
(I p)
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
Henrik Strahl, Leendert W Hamoen
Membrane potential is important for bacterial cell division.
Proc Natl Acad Sci U S A: 2010, 107(27);12281-6
[PubMed:20566861]
[WorldCat.org]
[DOI]
(I p)
Suey van Baarle, Marc Bramkamp
The MinCDJ system in Bacillus subtilis prevents minicell formation by promoting divisome disassembly.
PLoS One: 2010, 5(3);e9850
[PubMed:20352045]
[WorldCat.org]
[DOI]
(I e)
Marc Bramkamp, Robyn Emmins, Louise Weston, Catriona Donovan, Richard A Daniel, Jeff Errington
A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD.
Mol Microbiol: 2008, 70(6);1556-69
[PubMed:19019154]
[WorldCat.org]
[DOI]
(I p)
Warawan Eiamphungporn, John D Helmann
The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses.
Mol Microbiol: 2008, 67(4);830-48
[PubMed:18179421]
[WorldCat.org]
[DOI]
(P p)
David E Anderson, Frederico J Gueiros-Filho, Harold P Erickson
Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins.
J Bacteriol: 2004, 186(17);5775-81
[PubMed:15317782]
[WorldCat.org]
[DOI]
(P p)
Sabine Autret, Jeffery Errington
A role for division-site-selection protein MinD in regulation of internucleoid jumping of Soj (ParA) protein in Bacillus subtilis.
Mol Microbiol: 2003, 47(1);159-69
[PubMed:12492861]
[WorldCat.org]
[DOI]
(P p)
Frederico J Gueiros-Filho, Richard Losick
A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ.
Genes Dev: 2002, 16(19);2544-56
[PubMed:12368265]
[WorldCat.org]
[DOI]
(P p)
M E Karoui, J Errington
Isolation and characterization of topological specificity mutants of minD in Bacillus subtilis.
Mol Microbiol: 2001, 42(5);1211-21
[PubMed:11886553]
[WorldCat.org]
[DOI]
(P p)
A L Marston, J Errington
Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC.
Mol Microbiol: 1999, 33(1);84-96
[PubMed:10411726]
[WorldCat.org]
[DOI]
(P p)
A L Marston, H B Thomaides, D H Edwards, M E Sharpe, J Errington
Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site.
Genes Dev: 1998, 12(21);3419-30
[PubMed:9808628]
[WorldCat.org]
[DOI]
(P p)
S Lee, C W Price
The minCD locus of Bacillus subtilis lacks the minE determinant that provides topological specificity to cell division.
Mol Microbiol: 1993, 7(4);601-10
[PubMed:8459776]
[WorldCat.org]
[DOI]
(P p)
P A Levin, P S Margolis, P Setlow, R Losick, D Sun
Identification of Bacillus subtilis genes for septum placement and shape determination.
J Bacteriol: 1992, 174(21);6717-28
[PubMed:1400224]
[WorldCat.org]
[DOI]
(P p)