Difference between revisions of "Pgm"
| Line 61: | Line 61: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU33910&redirect=T BSU33910] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/cggR-gapA-pgk-tpiA-pgm-eno.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/cggR-gapA-pgk-tpiA-pgm-eno.html] | ||
| Line 100: | Line 101: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU33910&redirect=T BSU33910] | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1EJJ 1EJJ] (Geobacillus stearothermophilus, complex with 3-phosphoglycerate), [http://www.rcsb.org/pdb/explore.do?structureId=1EQJ 1EQJ] (Geobacillus stearothermophilus, complex with 2-phosphoglycerate), ''Geobacillus stearothermophilus'', complex with 2-phosphoglycerate [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=16359 NCBI], ''Geobacillus stearothermophilus'', complex with 3-phosphoglycerate [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=15578 NCBI] | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1EJJ 1EJJ] (Geobacillus stearothermophilus, complex with 3-phosphoglycerate), [http://www.rcsb.org/pdb/explore.do?structureId=1EQJ 1EQJ] (Geobacillus stearothermophilus, complex with 2-phosphoglycerate), ''Geobacillus stearothermophilus'', complex with 2-phosphoglycerate [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=16359 NCBI], ''Geobacillus stearothermophilus'', complex with 3-phosphoglycerate [http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?Dopt=s&uid=15578 NCBI] | ||
Revision as of 14:47, 2 April 2014
- Description: phosphoglycerate mutase, glycolytic / gluconeogenic enzyme
| Gene name | pgm |
| Synonyms | gpmI |
| Essential | Yes (PubMed) |
| Product | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase |
| Function | enzyme in glycolysis / gluconeogenesis |
| Gene expression levels in SubtiExpress: pgm | |
| Interactions involving this protein in SubtInteract: Pgm | |
| Metabolic function and regulation of this protein in SubtiPathways: pgm | |
| MW, pI | 56,1 kDa, 5.21 |
| Gene length, protein length | 1533 bp, 511 amino acids |
| Immediate neighbours | eno, tpi |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 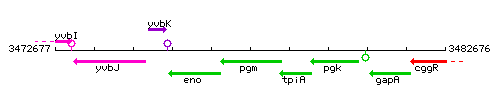
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed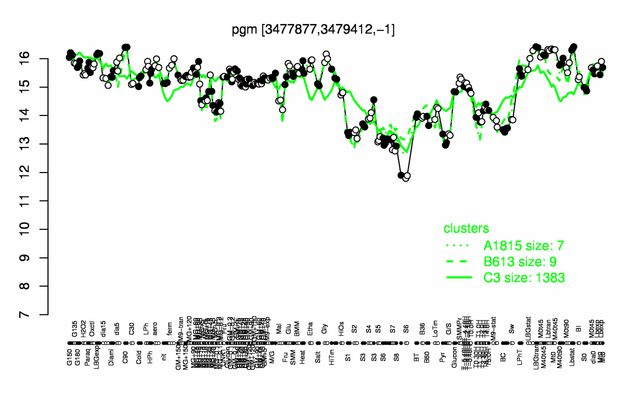
| |
Contents
Categories containing this gene/protein
carbon core metabolism, essential genes, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33910
Phenotypes of a mutant
- Essential PubMed
Database entries
- BsubCyc: BSU33910
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2-phospho-D-glycerate = 3-phospho-D-glycerate (according to Swiss-Prot)
- Protein family: BPG-independent phosphoglycerate mutase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information: Reversible Michaelis-Menten PubMed
- Domains:
- Cofactor(s): Mn2+
- Effectors of protein activity:
- Interactions:
- Pgm-PfkA
- Localization: Cytoplasm (Homogeneous) PubMed
Database entries
- BsubCyc: BSU33910
- Structure: 1EJJ (Geobacillus stearothermophilus, complex with 3-phosphoglycerate), 1EQJ (Geobacillus stearothermophilus, complex with 2-phosphoglycerate), Geobacillus stearothermophilus, complex with 2-phosphoglycerate NCBI, Geobacillus stearothermophilus, complex with 3-phosphoglycerate NCBI
- UniProt: P39773
- KEGG entry: [3]
- E.C. number: 5.4.2.1]
Additional information
- extensive information on the structure and enzymatic properties of Pgm can be found at Proteopedia
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- GP593 (pgm::cat), available in Jörg Stülke's lab, PubMed
- GP598 (pgm::erm), available in Jörg Stülke's lab, PubMed
- GP698 (pgm-eno::cat), available in Jörg Stülke's lab, PubMed
- Expression vector:
- pGP1425 (expression of pgm in B. subtilis, in pBQ200), available in Jörg Stülke's lab
- pGP1500 (expression of pgm and eno in B. subtilis, in pBQ200), available in Jörg Stülke's lab
- pGP1101 (N-terminal His-tag, in pWH844), available in Jörg Stülke's lab
- pGP396 (Pgm-S62A, N-terminal His-tag, in pWH844), available in Jörg Stülke's lab
- pGP92 (N-terminal Strep-tag, for SPINE, expression in B. subtilis, in pGP380), available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Mark J. Jedrzejas, Research Center Oakland, CA, USA Homepage
Your additional remarks
References
Fabian M Commichau, Nico Pietack, Jörg Stülke
Essential genes in Bacillus subtilis: a re-evaluation after ten years.
Mol Biosyst: 2013, 9(6);1068-75
[PubMed:23420519]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Laurent Jannière, Danielle Canceill, Catherine Suski, Sophie Kanga, Bérengère Dalmais, Roxane Lestini, Anne-Françoise Monnier, Jérôme Chapuis, Alexander Bolotin, Marina Titok, Emmanuelle Le Chatelier, S Dusko Ehrlich
Genetic evidence for a link between glycolysis and DNA replication.
PLoS One: 2007, 2(5);e447
[PubMed:17505547]
[WorldCat.org]
[DOI]
(I e)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Masatoshi Nukui, Luciane V Mello, James E Littlejohn, Barbara Setlow, Peter Setlow, Kijeong Kim, Terrance Leighton, Mark J Jedrzejas
Structure and molecular mechanism of Bacillus anthracis cofactor-independent phosphoglycerate mutase: a crucial enzyme for spores and growing cells of Bacillus species.
Biophys J: 2007, 92(3);977-88
[PubMed:17085493]
[WorldCat.org]
[DOI]
(P p)
Jean-Christophe Meile, Ling Juan Wu, S Dusko Ehrlich, Jeff Errington, Philippe Noirot
Systematic localisation of proteins fused to the green fluorescent protein in Bacillus subtilis: identification of new proteins at the DNA replication factory.
Proteomics: 2006, 6(7);2135-46
[PubMed:16479537]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
Daniel J Rigden, Ejvis Lamani, Luciane V Mello, James E Littlejohn, Mark J Jedrzejas
Insights into the catalytic mechanism of cofactor-independent phosphoglycerate mutase from X-ray crystallography, simulated dynamics and molecular modeling.
J Mol Biol: 2003, 328(4);909-20
[PubMed:12729763]
[WorldCat.org]
[DOI]
(P p)
Daniel J Rigden, Luciane V Mello, Peter Setlow, Mark J Jedrzejas
Structure and mechanism of action of a cofactor-dependent phosphoglycerate mutase homolog from Bacillus stearothermophilus with broad specificity phosphatase activity.
J Mol Biol: 2002, 315(5);1129-43
[PubMed:11827481]
[WorldCat.org]
[DOI]
(P p)
M J Jedrzejas, P Setlow
Comparison of the binuclear metalloenzymes diphosphoglycerate-independent phosphoglycerate mutase and alkaline phosphatase: their mechanism of catalysis via a phosphoserine intermediate.
Chem Rev: 2001, 101(3);607-18
[PubMed:11712498]
[WorldCat.org]
[DOI]
(P p)
D J Rigden, I Bagyan, E Lamani, P Setlow, M J Jedrzejas
A cofactor-dependent phosphoglycerate mutase homolog from Bacillus stearothermophilus is actually a broad specificity phosphatase.
Protein Sci: 2001, 10(9);1835-46
[PubMed:11514674]
[WorldCat.org]
[DOI]
(P p)
H Ludwig, G Homuth, M Schmalisch, F M Dyka, M Hecker, J Stülke
Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon.
Mol Microbiol: 2001, 41(2);409-22
[PubMed:11489127]
[WorldCat.org]
[DOI]
(P p)
M J Jedrzejas, M Chander, P Setlow, G Krishnasamy
Mechanism of catalysis of the cofactor-independent phosphoglycerate mutase from Bacillus stearothermophilus. Crystal structure of the complex with 2-phosphoglycerate.
J Biol Chem: 2000, 275(30);23146-53
[PubMed:10764795]
[WorldCat.org]
[DOI]
(P p)
M J Jedrzejas, M Chander, P Setlow, G Krishnasamy
Structure and mechanism of action of a novel phosphoglycerate mutase from Bacillus stearothermophilus.
EMBO J: 2000, 19(7);1419-31
[PubMed:10747010]
[WorldCat.org]
[DOI]
(P p)
M Chander, P Setlow, E Lamani, M J Jedrzejas
Structural studies on a 2,3-diphosphoglycerate independent phosphoglycerate mutase from Bacillus stearothermophilus.
J Struct Biol: 1999, 126(2);156-65
[PubMed:10388626]
[WorldCat.org]
[DOI]
(P p)
M Chander, B Setlow, P Setlow
The enzymatic activity of phosphoglycerate mutase from gram-positive endospore-forming bacteria requires Mn2+ and is pH sensitive.
Can J Microbiol: 1998, 44(8);759-67
[PubMed:9830105]
[WorldCat.org]
[DOI]
(P p)
N G Magill, A E Cowan, M A Leyva-Vazquez, M Brown, D E Koppel, P Setlow
Analysis of the relationship between the decrease in pH and accumulation of 3-phosphoglyceric acid in developing forespores of Bacillus species.
J Bacteriol: 1996, 178(8);2204-10
[PubMed:8636019]
[WorldCat.org]
[DOI]
(P p)
M A Leyva-Vazquez, P Setlow
Cloning and nucleotide sequences of the genes encoding triose phosphate isomerase, phosphoglycerate mutase, and enolase from Bacillus subtilis.
J Bacteriol: 1994, 176(13);3903-10
[PubMed:8021172]
[WorldCat.org]
[DOI]
(P p)
N J Kuhn, B Setlow, P Setlow
Manganese(II) activation of 3-phosphoglycerate mutase of Bacillus megaterium: pH-sensitive interconversion of active and inactive forms.
Arch Biochem Biophys: 1993, 306(2);342-9
[PubMed:8215434]
[WorldCat.org]
[DOI]
(P p)
K Watabe, E Freese
Purification and properties of the manganese-dependent phosphoglycerate mutase of Bacillus subtilis.
J Bacteriol: 1979, 137(2);773-8
[PubMed:33963]
[WorldCat.org]
[DOI]
(P p)