Difference between revisions of "Pyk"
| Line 79: | Line 79: | ||
=== Extended information on the protein === | === Extended information on the protein === | ||
| − | * '''Kinetic information:''' | + | * '''Kinetic information:''' |
| + | ** allosteric Regulation [http://www.ncbi.nlm.nih.gov/pubmed/3711058 PubMed] | ||
| + | ** specific activity: 0.231 µmol min<sup>-1</sup> (mg protein)<sup>-1</sup> {{PubMed|24571712}} | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' phosphorylation on Ser36 [http://www.ncbi.nlm.nih.gov/pubmed/17218307 PubMed], [http://www.ncbi.nlm.nih.gov/pubmed/16493705 PubMed], phosphorylation on Ser536 or Ser546 [http://www.ncbi.nlm.nih.gov/pubmed/17726680 PubMed], please note that the Ser is not on position 536 but rather at 538 | * '''Modification:''' phosphorylation on Ser36 [http://www.ncbi.nlm.nih.gov/pubmed/17218307 PubMed], [http://www.ncbi.nlm.nih.gov/pubmed/16493705 PubMed], phosphorylation on Ser536 or Ser546 [http://www.ncbi.nlm.nih.gov/pubmed/17726680 PubMed], please note that the Ser is not on position 536 but rather at 538 | ||
| − | * ''' | + | * '''[[Cofactors]]:''' Mg2+, K+ |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 153: | Line 155: | ||
=References= | =References= | ||
| − | <pubmed>17726680 16493705 11489127 17505547 10966427 17218307 3711058 4623707 21821766 22846916 23420519 24158146 </pubmed> | + | <pubmed>17726680 16493705 11489127 17505547 10966427 17218307 3711058 4623707 21821766 22846916 23420519 24158146 24571712</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 16:32, 28 February 2014
- Description: pyruvate kinase, glycolytic enzyme
| Gene name | pyk |
| Synonyms | pykA |
| Essential | no |
| Product | pyruvate kinase |
| Function | catabolic enzyme in glycolysis |
| Gene expression levels in SubtiExpress: pyk | |
| Metabolic function and regulation of this protein in SubtiPathways: pyk | |
| MW, pI | 61,9 kDa, 4.88 |
| Gene length, protein length | 1755 bp, 585 amino acids |
| Immediate neighbours | ytzA, pfkA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 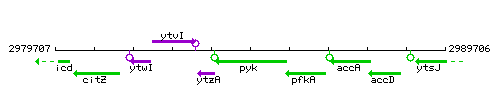
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
ATP synthesis, carbon core metabolism, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29180
Phenotypes of a mutant
Unable to grow with non-PTS carbohydrates (such as glucitol or glycerol) as single carbon source.
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ADP + phosphoenolpyruvate --> ATP + pyruvate
- The reaction is irreversible under physiological conditions
- Protein family: PEP-utilizing enzyme family (according to Swiss-Prot) pyruvate kinase family, (C-terminal section: PEP-utilizing enzyme family)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification: phosphorylation on Ser36 PubMed, PubMed, phosphorylation on Ser536 or Ser546 PubMed, please note that the Ser is not on position 536 but rather at 538
- Cofactors: Mg2+, K+
- Effectors of protein activity:
- Localization: cytoplasm PubMed
Database entries
- Structure: 2E28 (Geobacillus stearothermophilus)
- UniProt: P80885
- KEGG entry: [3]
- E.C. number: 2.7.1.40
Additional information
The enzyme is a tetramer with four active sites PubMed
Expression and regulation
- Regulation:
- twofold induced by glucose PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- GP589 (pyk::cat), available in Jörg Stülke's lab, PubMed
- GP600 (pyk::erm), available in Jörg Stülke's lab, PubMed
- Expression vector:
- expression in E. coli, N-terminal His-tag: pGP1100 (in pWH844), available in Jörg Stülke's lab
- expression in B. subtilis, native protein: pGP1411 (in pBQ200), available in Jörg Stülke's lab
- expression in B. subtilis, N-terminal Strep-tag: pGP1409 (in pGP380), available in Jörg Stülke's lab
- expression in B. subtilis, C-terminal Strep-tag: pGP1410 (in pGP382), available in Jörg Stülke's lab
- lacZ fusion: see pfkA
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Jörg Stülke's lab
- Antibody:
Labs working on this gene/protein
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References
Michael Kohlstedt, Praveen K Sappa, Hanna Meyer, Sandra Maaß, Adrienne Zaprasis, Tamara Hoffmann, Judith Becker, Leif Steil, Michael Hecker, Jan Maarten van Dijl, Michael Lalk, Ulrike Mäder, Jörg Stülke, Erhard Bremer, Uwe Völker, Christoph Wittmann
Adaptation of Bacillus subtilis carbon core metabolism to simultaneous nutrient limitation and osmotic challenge: a multi-omics perspective.
Environ Microbiol: 2014, 16(6);1898-917
[PubMed:24571712]
[WorldCat.org]
[DOI]
(I p)
Cuauhtemoc Licona-Cassani, Alvaro R Lara, Natividad Cabrera-Valladares, Adelfo Escalante, Georgina Hernández-Chávez, Alfredo Martinez, Francisco Bolívar, Guillermo Gosset
Inactivation of pyruvate kinase or the phosphoenolpyruvate: sugar phosphotransferase system increases shikimic and dehydroshikimic acid yields from glucose in Bacillus subtilis.
J Mol Microbiol Biotechnol: 2014, 24(1);37-45
[PubMed:24158146]
[WorldCat.org]
[DOI]
(I p)
Fabian M Commichau, Nico Pietack, Jörg Stülke
Essential genes in Bacillus subtilis: a re-evaluation after ten years.
Mol Biosyst: 2013, 9(6);1068-75
[PubMed:23420519]
[WorldCat.org]
[DOI]
(I p)
Natividad Cabrera-Valladares, Luz M Martínez, Noemí Flores, Georgina Hernández-Chávez, Alfredo Martínez, Francisco Bolívar, Guillermo Gosset
Physiologic consequences of glucose transport and phosphoenolpyruvate node modifications in Bacillus subtilis 168.
J Mol Microbiol Biotechnol: 2012, 22(3);177-97
[PubMed:22846916]
[WorldCat.org]
[DOI]
(I p)
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Dörte Becher, Jörg Bernhardt, Katrin Gronau, Anja Klutzny, Michael Hecker
Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis.
Proteomics: 2007, 7(19);3509-26
[PubMed:17726680]
[WorldCat.org]
[DOI]
(P p)
Laurent Jannière, Danielle Canceill, Catherine Suski, Sophie Kanga, Bérengère Dalmais, Roxane Lestini, Anne-Françoise Monnier, Jérôme Chapuis, Alexander Bolotin, Marina Titok, Emmanuelle Le Chatelier, S Dusko Ehrlich
Genetic evidence for a link between glycolysis and DNA replication.
PLoS One: 2007, 2(5);e447
[PubMed:17505547]
[WorldCat.org]
[DOI]
(I e)
Boris Macek, Ivan Mijakovic, Jesper V Olsen, Florian Gnad, Chanchal Kumar, Peter R Jensen, Matthias Mann
The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis.
Mol Cell Proteomics: 2007, 6(4);697-707
[PubMed:17218307]
[WorldCat.org]
[DOI]
(P p)
Alain Lévine, Françoise Vannier, Cédric Absalon, Lauriane Kuhn, Peter Jackson, Elaine Scrivener, Valérie Labas, Joëlle Vinh, Patrick Courtney, Jérôme Garin, Simone J Séror
Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes.
Proteomics: 2006, 6(7);2157-73
[PubMed:16493705]
[WorldCat.org]
[DOI]
(P p)
H Ludwig, G Homuth, M Schmalisch, F M Dyka, M Hecker, J Stülke
Transcription of glycolytic genes and operons in Bacillus subtilis: evidence for the presence of multiple levels of control of the gapA operon.
Mol Microbiol: 2001, 41(2);409-22
[PubMed:11489127]
[WorldCat.org]
[DOI]
(P p)
B Fry, T Zhu, M M Domach, R R Koepsel, C Phalakornkule, M M Ataai
Characterization of growth and acid formation in a Bacillus subtilis pyruvate kinase mutant.
Appl Environ Microbiol: 2000, 66(9);4045-9
[PubMed:10966427]
[WorldCat.org]
[DOI]
(P p)
H Sakai, K Suzuki, K Imahori
Purification and properties of pyruvate kinase from Bacillus stearothermophilus.
J Biochem: 1986, 99(4);1157-67
[PubMed:3711058]
[WorldCat.org]
[DOI]
(P p)
M Diesterhaft, E Freese
Pyruvate kinase of bacillus subtilis.
Biochim Biophys Acta: 1972, 268(2);373-80
[PubMed:4623707]
[WorldCat.org]
[DOI]
(P p)