Difference between revisions of "MtrB"
(→Original Publications) |
|||
| Line 64: | Line 64: | ||
=== Additional information=== | === Additional information=== | ||
| + | * mutations in ''[[mtrB]]'', ''[[sigB]]'', ''[[rpoB]]'', and ''[[rpoC]]'' allow ''B. subtilis'' to grow with 4-fluorotryptophan rather than with tryptophan as a canonical amino acid of the genetic code {{PubMed|24572018}} | ||
=The protein= | =The protein= | ||
Revision as of 16:20, 28 February 2014
- Description: tryptophan operon RNA-binding attenuation protein (TRAP), controls the RNA switch in front of genes involved in biosynthesis and acquisition of tryptophan
| Gene name | mtrB |
| Synonyms | |
| Essential | no |
| Product | tryptophan operon RNA-binding attenuation protein (TRAP) |
| Function | regulation of tryptophan biosynthesis (and translation) attenuation in the trp operon; repression of the folate operon |
| Gene expression levels in SubtiExpress: mtrB | |
| Interactions involving this protein in SubtInteract: TRAP | |
| Metabolic function and regulation of this protein in SubtiPathways: mtrB | |
| MW, pI | 8 kDa, 7.333 |
| Gene length, protein length | 225 bp, 75 aa |
| Immediate neighbours | hepS, folE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 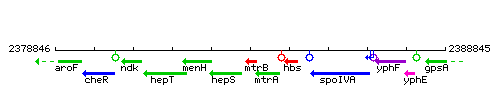
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids, transcription factors and their control, RNA binding regulators
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22770
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- mutations in mtrB, sigB, rpoB, and rpoC allow B. subtilis to grow with 4-fluorotryptophan rather than with tryptophan as a canonical amino acid of the genetic code PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: prpD family (according to Swiss-Prot)
- Paralogous protein(s):
Genes controlled by MtrB
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- binding of tryptophan results RNA binding and thus in transcription termination
Database entries
- UniProt: P19466
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original Publications