Difference between revisions of "CypC"
Raphael2215 (talk | contribs) (→Database entries) |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' long chain-fatty acid beta-hydroxylating [[cytochrome P450]], H(2)O(2)-dependent, hydroxylates myristic acid to beta-hydroxymyristic acid<br/><br/> | + | * '''Description:''' long chain-fatty acid beta-hydroxylating [[cytochrome P450]], H(2)O(2)-dependent, hydroxylates myristic acid to beta-hydroxymyristic acid, required for protection against paraquat stress <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || fatty acid beta-hydroxylating [[cytochrome P450]] | + | |style="background:#ABCDEF;" align="center"| '''Product''' || fatty acid beta-hydroxylating [[cytochrome P450]], protection against paraquat stress |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || biosynthesis of beta-hydroxy fatty acid for lipopeptides | |style="background:#ABCDEF;" align="center"|'''Function''' || biosynthesis of beta-hydroxy fatty acid for lipopeptides | ||
| Line 39: | Line 39: | ||
= [[Categories]] containing this gene/protein = | = [[Categories]] containing this gene/protein = | ||
{{SubtiWiki category|[[general stress proteins (controlled by SigB)]]}}, | {{SubtiWiki category|[[general stress proteins (controlled by SigB)]]}}, | ||
| + | {{SubtiWiki category|[[resistance against oxidative and electrophile stress]]}}, | ||
{{SubtiWiki category|[[electron transport/ other]]}}, | {{SubtiWiki category|[[electron transport/ other]]}}, | ||
{{SubtiWiki category|[[lipid metabolism/ other]]}} | {{SubtiWiki category|[[lipid metabolism/ other]]}} | ||
| Line 136: | Line 137: | ||
=References= | =References= | ||
| − | <pubmed>12297285 11827534 11566026 17385817,11914497 10529095,12519760,15805528, 23586998 20697922,21673922</pubmed> | + | <pubmed>12297285 11827534 11566026 17385817,11914497 10529095,12519760,15805528, 23586998 20697922,21673922 22582280</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:13, 15 September 2013
- Description: long chain-fatty acid beta-hydroxylating cytochrome P450, H(2)O(2)-dependent, hydroxylates myristic acid to beta-hydroxymyristic acid, required for protection against paraquat stress
| Gene name | cypC |
| Synonyms | ybdT |
| Essential | no |
| Product | fatty acid beta-hydroxylating cytochrome P450, protection against paraquat stress |
| Function | biosynthesis of beta-hydroxy fatty acid for lipopeptides |
| Gene expression levels in SubtiExpress: cypC | |
| MW, pI | 47 kDa, 6.468 |
| Gene length, protein length | 1251 bp, 417 aa |
| Immediate neighbours | ybxI, ybyB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 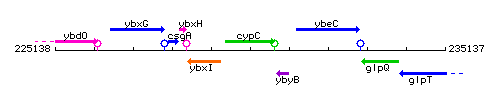
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
general stress proteins (controlled by SigB), resistance against oxidative and electrophile stress, electron transport/ other, lipid metabolism/ other
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU02100
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- hydroxylation of a long-chain fatty acid (e.g. myristic acid) at the alpha- and beta-positions using hydrogen peroxide as an oxidant PubMed
- Protein family: cytochrome P450 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: O31440
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: cypC (according to DBTBS)
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References