Difference between revisions of "MutM"
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 111: | Line 107: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=mutM_2972329_2973159_-1 mutM] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=mutM_2972329_2973159_-1 mutM] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 138: | Line 134: | ||
=References= | =References= | ||
| + | ==Reviews== | ||
| + | <pubmed> 22933559 </pubmed> | ||
| + | == Original publications == | ||
<pubmed> 19011023 19889642 </pubmed> | <pubmed> 19011023 19889642 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 17:10, 13 September 2013
- Description: formamidopyrimidine-DNA glycosidase
| Gene name | mutM |
| Synonyms | ytaE |
| Essential | no |
| Product | formamidopyrimidine-DNA glycosidase |
| Function | DNA repair |
| Gene expression levels in SubtiExpress: mutM | |
| MW, pI | 31 kDa, 8.889 |
| Gene length, protein length | 834 bp, 278 aa |
| Immediate neighbours | ytaF, polA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 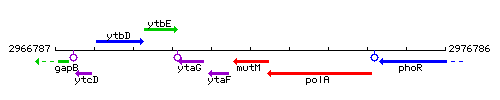
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed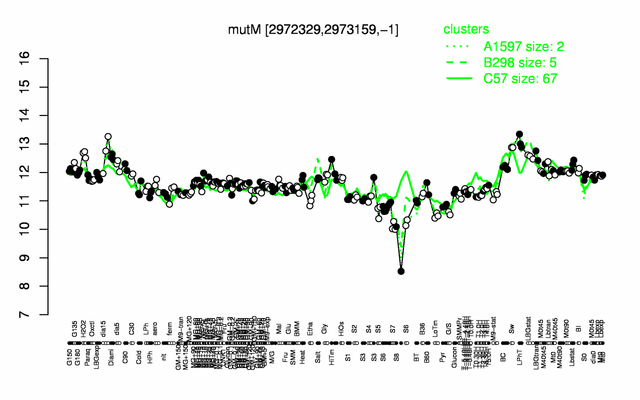
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29080
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolysis of DNA containing ring-opened 7-methylguanine residues, releasing 2,6-diamino-4-hydroxy-5-(N-methyl)formamidopyrimidine (according to Swiss-Prot)
- Protein family: FPG family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- Structure: 2F5S (complex with oxoG:C, Geobacillus stearothermophilus)
- UniProt: O34403
- KEGG entry: [2]
- E.C. number: 3.2.2.23
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Justin S Lenhart, Jeremy W Schroeder, Brian W Walsh, Lyle A Simmons
DNA repair and genome maintenance in Bacillus subtilis.
Microbiol Mol Biol Rev: 2012, 76(3);530-64
[PubMed:22933559]
[WorldCat.org]
[DOI]
(I p)
Original publications
Yan Qi, Marie C Spong, Kwangho Nam, Martin Karplus, Gregory L Verdine
Entrapment and structure of an extrahelical guanine attempting to enter the active site of a bacterial DNA glycosylase, MutM.
J Biol Chem: 2010, 285(2);1468-78
[PubMed:19889642]
[WorldCat.org]
[DOI]
(I p)
Luz E Vidales, Lluvia C Cárdenas, Eduardo Robleto, Ronald E Yasbin, Mario Pedraza-Reyes
Defects in the error prevention oxidized guanine system potentiate stationary-phase mutagenesis in Bacillus subtilis.
J Bacteriol: 2009, 191(2);506-13
[PubMed:19011023]
[WorldCat.org]
[DOI]
(I p)