Difference between revisions of "SpoIIIC"
| Line 30: | Line 30: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| − | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=spoIIIC_2701338_2701754_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:spoIIIC_expression.png|500px]] | + | |colspan="2" |'''[http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=spoIIIC_2701338_2701754_1 Expression at a glance]'''   {{PubMed|22383849}}<br/>[[Image:spoIIIC_expression.png|500px|link=http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU26390]] |
|- | |- | ||
|} | |} | ||
Revision as of 14:01, 16 May 2013
- Description: RNA polymerase sporulation-specific sigma factor (SigK) (C-terminal half), other part: spoIVCB
| Gene name | spoIIIC |
| Synonyms | sigK, spoIVD, spoIVE |
| Essential | no |
| Product | RNA polymerase sporulation-specific sigma factor (SigK) (C-terminal half) |
| Function | late mother cell-specific gene expression |
| Gene expression levels in SubtiExpress: spoIIIC | |
| Interactions involving this protein in SubtInteract: SpoIIIC | |
| MW, pI | 16 kDa, 9.608 |
| Gene length, protein length | 414 bp, 138 aa |
| Immediate neighbours | yqaB, yrkS |
| Gene sequence (+200bp) | Protein sequence |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed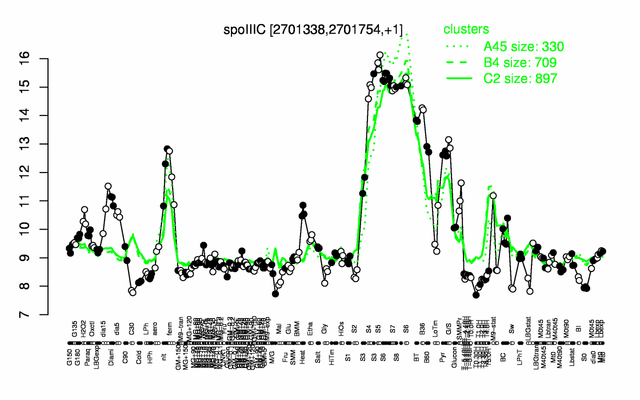
| |
Contents
Categories containing this gene/protein
transcription, sigma factors and their control, sporulation proteins, membrane proteins
This gene is a member of the following regulons
GerE regulon, SigE regulon, SigK regulon, SpoIIID regulon
The SigK regulon
The gene
Basic information
- Locus tag: BSU26390
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P12254
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
The SigK regulon
Jessica M Silvaggi, John B Perkins, Richard Losick
Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis.
J Bacteriol: 2006, 188(2);532-41
[PubMed:16385044]
[WorldCat.org]
[DOI]
(P p)
Patrick Eichenberger, Masaya Fujita, Shane T Jensen, Erin M Conlon, David Z Rudner, Stephanie T Wang, Caitlin Ferguson, Koki Haga, Tsutomu Sato, Jun S Liu, Richard Losick
The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis.
PLoS Biol: 2004, 2(10);e328
[PubMed:15383836]
[WorldCat.org]
[DOI]
(I p)
Other original publications