Difference between revisions of "LicT"
| Line 26: | Line 26: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[bglS]]'', ''[[yxiP]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[bglS]]'', ''[[yxiP]]'' | ||
|- | |- | ||
| − | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU39080 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU39080 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU39080 | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU39080 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU39080 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU39080 DNA_with_flanks] |
|- | |- | ||
|- | |- | ||
Revision as of 11:39, 14 May 2013
- Description: transcriptional antiterminator of the bglP-bglH-yxiE operon and the bglS gene
| Gene name | licT |
| Synonyms | |
| Essential | no |
| Product | transcriptional antiterminator (BglG family) |
| Function | control of beta-glucan and beta-glucoside utilization |
| Gene expression levels in SubtiExpress: licT | |
| Interactions involving this protein in SubtInteract: LicT | |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 32 kDa, 5.944 |
| Gene length, protein length | 831 bp, 277 aa |
| Immediate neighbours | bglS, yxiP |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 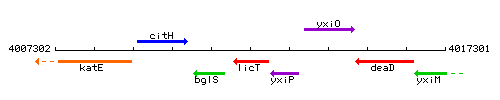
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed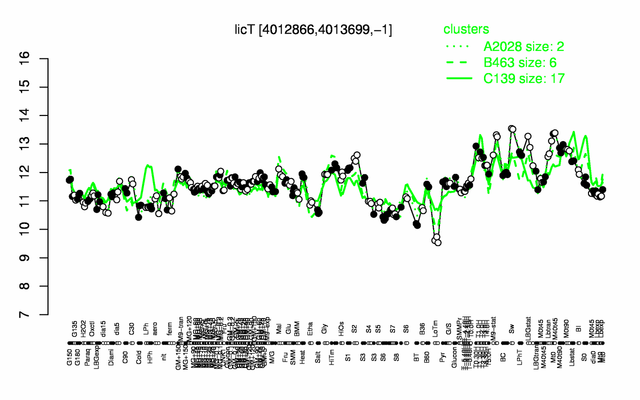
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, transcription factors and their control, RNA binding regulators, phosphoproteins
This gene is a member of the following regulons
The LicT regulon: bglP-bglH-yxiE, bglS
The gene
Basic information
- Locus tag: BSU39080
Phenotypes of a mutant
no expression of the bglP-bglH operon
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: binding to the mRNAs of bglS and the bglP-bglH operon, causes transcription antitermination (in presence of salicin and absence of glucose)
- Protein family: transcriptional antiterminator BglG family of antiterminators (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- K(D) for the RAT-RNA: 10 nM PubMed
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm, even distribution in the absence of the inducer salicin, subpolar localization in the presence of salicin PubMed
Database entries
- UniProt: P39805
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP427 (licTS, erm), available in the Stülke lab
- Expression vector:
- for expression, purification of both PRDs in E. coli with N-terminal His-tag, in pWH844: pGP165, available in Stülke lab
- for expression, purification of the RNA-binding domain in E. coli with N-terminal His-tag, in pWH844: pGP315, available in Stülke lab
- for expression, purification of the RNA-binding domain in E. coli with N-terminal His-tag and thrombin cleavage site, in pGP570: pGP572, available in Stülke lab
- lacZ fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Stephane Aymerich, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Josef Deutscher, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Michael Hecker, Greifswald, Germany Homepage
Your additional remarks
References
Original description
Control of LicT activity
Fabian M Rothe, Christoph Wrede, Martin Lehnik-Habrink, Boris Görke, Jörg Stülke
Dynamic localization of a transcription factor in Bacillus subtilis: the LicT antiterminator relocalizes in response to inducer availability.
J Bacteriol: 2013, 195(10);2146-54
[PubMed:23475962]
[WorldCat.org]
[DOI]
(I p)
Thomas Bahr, Denise Lüttmann, Walter März, Bodo Rak, Boris Görke
Insight into bacterial phosphotransferase system-mediated signaling by interspecies transplantation of a transcriptional regulator.
J Bacteriol: 2011, 193(8);2013-26
[PubMed:21335451]
[WorldCat.org]
[DOI]
(I p)
Cordula Lindner, Michael Hecker, Dominique Le Coq, Josef Deutscher
Bacillus subtilis mutant LicT antiterminators exhibiting enzyme I- and HPr-independent antitermination affect catabolite repression of the bglPH operon.
J Bacteriol: 2002, 184(17);4819-28
[PubMed:12169607]
[WorldCat.org]
[DOI]
(P p)
P Tortosa, N Declerck, H Dutartre, C Lindner, J Deutscher, D Le Coq
Sites of positive and negative regulation in the Bacillus subtilis antiterminators LicT and SacY.
Mol Microbiol: 2001, 41(6);1381-93
[PubMed:11580842]
[WorldCat.org]
[DOI]
(P p)
C Lindner, A Galinier, M Hecker, J Deutscher
Regulation of the activity of the Bacillus subtilis antiterminator LicT by multiple PEP-dependent, enzyme I- and HPr-catalysed phosphorylation.
Mol Microbiol: 1999, 31(3);995-1006
[PubMed:10048041]
[WorldCat.org]
[DOI]
(P p)
S Krüger, S Gertz, M Hecker
Transcriptional analysis of bglPH expression in Bacillus subtilis: evidence for two distinct pathways mediating carbon catabolite repression.
J Bacteriol: 1996, 178(9);2637-44
[PubMed:8626332]
[WorldCat.org]
[DOI]
(P p)
Structural analysis of LicT
Hélène Déméné, Thierry Ducat, Karine De Guillen, Catherine Birck, Stéphane Aymerich, Michel Kochoyan, Nathalie Declerck
Structural mechanism of signal transduction between the RNA-binding domain and the phosphotransferase system regulation domain of the LicT antiterminator.
J Biol Chem: 2008, 283(45);30838-49
[PubMed:18682383]
[WorldCat.org]
[DOI]
(P p)
Marc Graille, Cong-Zhao Zhou, Véronique Receveur-Bréchot, Bruno Collinet, Nathalie Declerck, Herman van Tilbeurgh
Activation of the LicT transcriptional antiterminator involves a domain swing/lock mechanism provoking massive structural changes.
J Biol Chem: 2005, 280(15);14780-9
[PubMed:15699035]
[WorldCat.org]
[DOI]
(P p)
N Declerck, H Dutartre, V Receveur, V Dubois, C Royer, S Aymerich, H van Tilbeurgh
Dimer stabilization upon activation of the transcriptional antiterminator LicT.
J Mol Biol: 2001, 314(4);671-81
[PubMed:11733988]
[WorldCat.org]
[DOI]
(P p)
H van Tilbeurgh, D Le Coq, N Declerck
Crystal structure of an activated form of the PTS regulation domain from the LicT transcriptional antiterminator.
EMBO J: 2001, 20(14);3789-99
[PubMed:11447120]
[WorldCat.org]
[DOI]
(P p)
LicT-RNA interaction
Caroline Clerte, Nathalie Declerck, Emmanuel Margeat
Competitive folding of anti-terminator/terminator hairpins monitored by single molecule FRET.
Nucleic Acids Res: 2013, 41(4);2632-43
[PubMed:23303779]
[WorldCat.org]
[DOI]
(I p)
Sebastian Hübner, Nathalie Declerck, Christine Diethmaier, Dominique Le Coq, Stephane Aymerich, Jörg Stülke
Prevention of cross-talk in conserved regulatory systems: identification of specificity determinants in RNA-binding anti-termination proteins of the BglG family.
Nucleic Acids Res: 2011, 39(10);4360-72
[PubMed:21278164]
[WorldCat.org]
[DOI]
(I p)
Hélène Déméné, Thierry Ducat, Karine De Guillen, Catherine Birck, Stéphane Aymerich, Michel Kochoyan, Nathalie Declerck
Structural mechanism of signal transduction between the RNA-binding domain and the phosphotransferase system regulation domain of the LicT antiterminator.
J Biol Chem: 2008, 283(45);30838-49
[PubMed:18682383]
[WorldCat.org]
[DOI]
(P p)
Yinshan Yang, Nathalie Declerck, Xavier Manival, Stéphane Aymerich, Michel Kochoyan
Solution structure of the LicT-RNA antitermination complex: CAT clamping RAT.
EMBO J: 2002, 21(8);1987-97
[PubMed:11953318]
[WorldCat.org]
[DOI]
(P p)
N Declerck, F Vincent, F Hoh, S Aymerich, H van Tilbeurgh
RNA recognition by transcriptional antiterminators of the BglG/SacY family: functional and structural comparison of the CAT domain from SacY and LicT.
J Mol Biol: 1999, 294(2);389-402
[PubMed:10610766]
[WorldCat.org]
[DOI]
(P p)
S Aymerich, M Steinmetz
Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family.
Proc Natl Acad Sci U S A: 1992, 89(21);10410-4
[PubMed:1279678]
[WorldCat.org]
[DOI]
(P p)