Difference between revisions of "ManR"
(→References) |
|||
| Line 13: | Line 13: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || regulation of mannose utilization | |style="background:#ABCDEF;" align="center"|'''Function''' || regulation of mannose utilization | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http:// | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU12000 manR] |
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/ManR ManR] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/ManR ManR] | ||
| Line 25: | Line 25: | ||
|style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yjdB]]'', ''[[manP]]'' | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[yjdB]]'', ''[[manP]]'' | ||
|- | |- | ||
| − | | | + | |style="background:#FAF8CC;" align="center"|'''Sequences'''||[http://bsubcyc.org/BSUB/sequence-aa?type=GENE&object=BSU12000 Protein] [http://bsubcyc.org/BSUB/sequence?type=GENE&object=BSU12000 DNA] [http://bsubcyc.org/BSUB/seq-selector?chromosome=CHROM-1&object=BSU12000 Advanced_DNA] |
|- | |- | ||
|- | |- | ||
Revision as of 12:34, 13 May 2013
| Gene name | manR |
| Synonyms | yjdC |
| Essential | no |
| Product | transcriptional activator, PRD-type |
| Function | regulation of mannose utilization |
| Gene expression levels in SubtiExpress: manR | |
| Interactions involving this protein in SubtInteract: ManR | |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 75 kDa, 8.04 |
| Gene length, protein length | 1944 bp, 648 aa |
| Immediate neighbours | yjdB, manP |
| Sequences | Protein DNA Advanced_DNA |
Genetic context 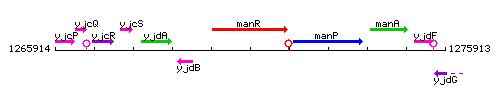
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed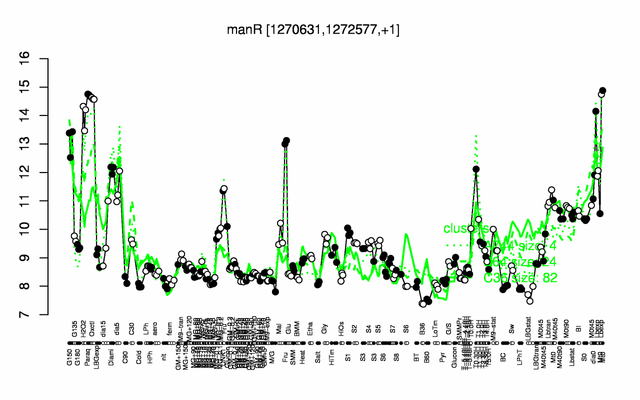
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources, transcription factors and their control, phosphoproteins
This gene is a member of the following regulons
The ManR regulon:
The gene
Basic information
- Locus tag: BSU12000
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: phosphorylation by ManP inactivates ManR (in the absence of mannose), phosphorylation by HPr stimluates ManR PubMed
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: O31644
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Marian Wenzel, Josef Altenbuchner
The Bacillus subtilis mannose regulator, ManR, a DNA-binding protein regulated by HPr and its cognate PTS transporter ManP.
Mol Microbiol: 2013, 88(3);562-76
[PubMed:23551403]
[WorldCat.org]
[DOI]
(I p)
Bogumiła C Marciniak, Monika Pabijaniak, Anne de Jong, Robert Dűhring, Gerald Seidel, Wolfgang Hillen, Oscar P Kuipers
High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis.
BMC Genomics: 2012, 13;401
[PubMed:22900538]
[WorldCat.org]
[DOI]
(I e)
Tianqi Sun, Josef Altenbuchner
Characterization of a mannose utilization system in Bacillus subtilis.
J Bacteriol: 2010, 192(8);2128-39
[PubMed:20139185]
[WorldCat.org]
[DOI]
(I p)
Jonathan Reizer, Steffi Bachem, Aiala Reizer, Maryvonne Arnaud, Milton H Saier, Jörg Stülke
Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 12);3419-3429
[PubMed:10627040]
[WorldCat.org]
[DOI]
(P p)
J Stülke, M Arnaud, G Rapoport, I Martin-Verstraete
PRD--a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria.
Mol Microbiol: 1998, 28(5);865-74
[PubMed:9663674]
[WorldCat.org]
[DOI]
(P p)