Difference between revisions of "DnaG"
(→Expression and regulation) |
|||
| Line 89: | Line 89: | ||
* '''Cofactor(s):''' | * '''Cofactor(s):''' | ||
| − | * '''Effectors of protein activity:''' the enzymatic activity is inhibited by (p)ppGpp during the ´stringent response´ {{PubMed|17350574}} | + | * '''Effectors of protein activity:''' |
| + | ** the enzymatic activity is inhibited by (p)ppGpp during the ´stringent response´ {{PubMed|17350574}} | ||
| + | ** [[DnaG]] primase activity is stimulates by [[DnaC]] {{PubMed|23563155}} | ||
* '''[[SubtInteract|Interactions]]:''' | * '''[[SubtInteract|Interactions]]:''' | ||
** part of the [[replisome]]: [[PolC]]-[[HolA]]-[[HolB]]-[[DnaX]]-[[DnaN]]-[[DnaG]]-[[DnaC]]-[[DnaI]]-[[DnaD]]-[[SsbA]]-[[DnaE]]-[[PriA]]-[[DnaB]] {{PubMed|20122408}} | ** part of the [[replisome]]: [[PolC]]-[[HolA]]-[[HolB]]-[[DnaX]]-[[DnaN]]-[[DnaG]]-[[DnaC]]-[[DnaI]]-[[DnaD]]-[[SsbA]]-[[DnaE]]-[[PriA]]-[[DnaB]] {{PubMed|20122408}} | ||
** part of the [[primosome]]: [[DnaA]]-[[DnaG]]-[[DnaC]]-[[DnaI]]-[[DnaD]]-[[DnaB]] {{PubMed|22797751}} | ** part of the [[primosome]]: [[DnaA]]-[[DnaG]]-[[DnaC]]-[[DnaI]]-[[DnaD]]-[[DnaB]] {{PubMed|22797751}} | ||
| + | ** [[DnaE]]-[[DnaG]] {{PubMed|23563155}} | ||
| + | ** [[DnaG]]-[[DnaC]] {{PubMed|23563155}} | ||
* '''[[Localization]]:''' Cytoplasm (Homogeneous) [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] | * '''[[Localization]]:''' Cytoplasm (Homogeneous) [http://www.ncbi.nlm.nih.gov/sites/entrez/16479537 PubMed] | ||
| Line 150: | Line 154: | ||
==Original publications== | ==Original publications== | ||
| − | + | <pubmed>3919021, 20122408,14651647, 16479537, 19415803 17350574 23563155 23268446,21419346</pubmed> | |
| − | <pubmed>3919021, 20122408,14651647, 16479537, 19415803 17350574 | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:24, 15 April 2013
- Description: DNA primase, part of the replisome
| Gene name | dnaG |
| Synonyms | dnaE |
| Essential | yes PubMed |
| Product | DNA primase |
| Function | DNA replication |
| Gene expression levels in SubtiExpress: dnaG | |
| Interactions involving this protein in SubtInteract: DnaG | |
| MW, pI | 68 kDa, 6.706 |
| Gene length, protein length | 1809 bp, 603 aa |
| Immediate neighbours | sigA, antE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 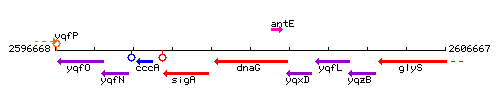
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed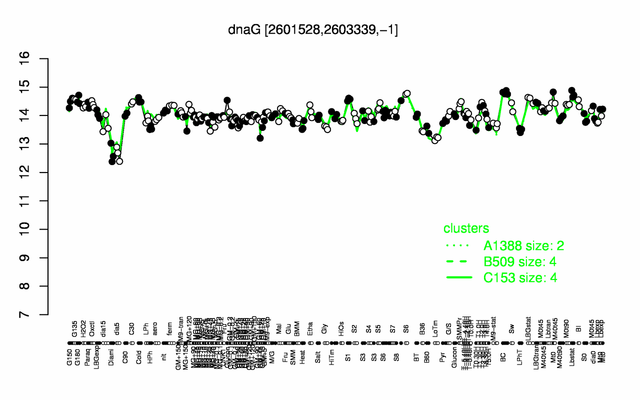
| |
Contents
Categories containing this gene/protein
DNA replication, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU25210
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- required for bacteriophage SPP1 replication PubMed
- Protein family: DNA primase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: Cytoplasm (Homogeneous) PubMed
Database entries
- Structure: 1D0Q (zinc binding domain, Geobacillus stearothermophilus), 1Z8S (DnaB binding domain, AA 452-597, Geobacillus stearothermophilus)
- UniProt: P05096
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
- enzymatic activity is inhibited by (p)ppGpp during the ´stringent response´ PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications