Difference between revisions of "BglP"
(→References) |
(→References) |
||
| Line 160: | Line 160: | ||
[http://www.ncbi.nlm.nih.gov/pubmed/23475962 PubMed:23475962] | [http://www.ncbi.nlm.nih.gov/pubmed/23475962 PubMed:23475962] | ||
<pubmed>16672620,20525796,10627040</pubmed> | <pubmed>16672620,20525796,10627040</pubmed> | ||
| − | <pubmed> | + | <pubmed>7559347, 22900538</pubmed> |
| + | <pubmed>7883710 8626332</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 09:50, 14 March 2013
- Description: trigger enzyme: beta-glucoside-specific phosphotransferase system, EIIBCA of the PTS
| Gene name | bglP |
| Synonyms | sytA |
| Essential | no |
| Product | trigger enzyme: beta-glucoside-specific phosphotransferase system, EIIBCA |
| Function | beta-glucoside uptake and phosphorylation, control of LicT activity |
| Gene expression levels in SubtiExpress: bglP | |
| Interactions involving this protein in SubtInteract: BglP | |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 64 kDa, 6.809 |
| Gene length, protein length | 1827 bp, 609 aa |
| Immediate neighbours | bglH, yxxE |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 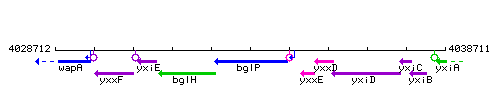
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed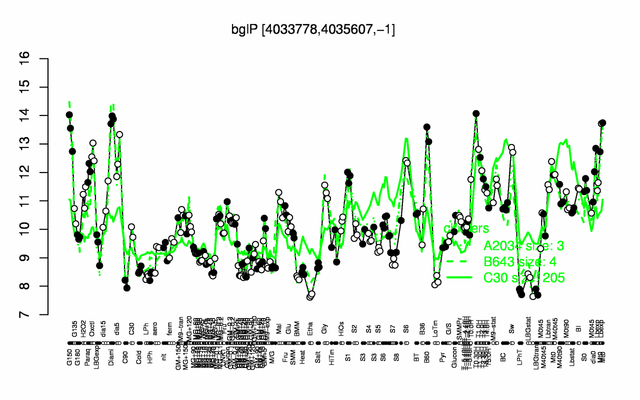
| |
Contents
Categories containing this gene/protein
phosphotransferase systems, utilization of specific carbon sources, transcription factors and their control, trigger enzyme, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU39270
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine/cysteine (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cell membrane PubMed
Database entries
- Structure:
- UniProt: P40739
- KEGG entry: [3]
- E.C. number: 2.7.1.69 9
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
- An antisense RNA is predicted for bglP PubMed
Biological materials
- Mutant: GP475 (erm), available in the Stülke lab
- Expression vector:
- GFP fusion:
- CFP fusion: B. subtilis GP1266 bglP-cfp ermC- without terminator, available in Jörg Stülke's lab
- YFP fusion: B. subtilis GP1274 bglP-yfp ermC- without terminator, available in Jörg Stülke's lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Fabian M Rothe, Christoph Wrede, Martin Lehnik-Habrink, Boris Görke, Jörg Stülke Dynamic localization of a transcription factor in Bacillus subtilis: The LicT antiterminator relocalizes in response to inducer availability. J Bacteriol.: 2013, 195, Epub ahead of print. PubMed:23475962
Irnov Irnov, Cynthia M Sharma, Jörg Vogel, Wade C Winkler
Identification of regulatory RNAs in Bacillus subtilis.
Nucleic Acids Res: 2010, 38(19);6637-51
[PubMed:20525796]
[WorldCat.org]
[DOI]
(I p)
Juliane Ollinger, Kyung-Bok Song, Haike Antelmann, Michael Hecker, John D Helmann
Role of the Fur regulon in iron transport in Bacillus subtilis.
J Bacteriol: 2006, 188(10);3664-73
[PubMed:16672620]
[WorldCat.org]
[DOI]
(P p)
Jonathan Reizer, Steffi Bachem, Aiala Reizer, Maryvonne Arnaud, Milton H Saier, Jörg Stülke
Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 12);3419-3429
[PubMed:10627040]
[WorldCat.org]
[DOI]
(P p)
Bogumiła C Marciniak, Monika Pabijaniak, Anne de Jong, Robert Dűhring, Gerald Seidel, Wolfgang Hillen, Oscar P Kuipers
High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis.
BMC Genomics: 2012, 13;401
[PubMed:22900538]
[WorldCat.org]
[DOI]
(I e)
S Krüger, M Hecker
Regulation of the putative bglPH operon for aryl-beta-glucoside utilization in Bacillus subtilis.
J Bacteriol: 1995, 177(19);5590-7
[PubMed:7559347]
[WorldCat.org]
[DOI]
(P p)