Difference between revisions of "RsbR"
(→Original Articles) |
|||
| Line 150: | Line 150: | ||
==Original Articles== | ==Original Articles== | ||
| − | '''Additional publications:''' {{PubMed|21362065,20935101,22287516}} | + | '''Additional publications:''' {{PubMed|21362065,20935101,22287516,22609918}} |
<pubmed>8002610,8682769,8682789, 17726680,10781545,15583165, 8824586,10329124,17158665, 9179850,8808936,15312768, 11244072,15342582,, 12950928, 15466036, 9179850, 8955331, 18832644 ,17726680 ,17218307 20019076 21602359 </pubmed> | <pubmed>8002610,8682769,8682789, 17726680,10781545,15583165, 8824586,10329124,17158665, 9179850,8808936,15312768, 11244072,15342582,, 12950928, 15466036, 9179850, 8955331, 18832644 ,17726680 ,17218307 20019076 21602359 </pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 10:59, 23 May 2012
- Description: activator of RsbT kinase activity, part of the stressosome
| Gene name | rsbR |
| Synonyms | ycxR, rsbRA |
| Essential | no |
| Product | activator of RsbT kinase activity |
| Function | control of SigB activity |
| Interactions involving this protein in SubtInteract: RbsR | |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 30 kDa, 4.731 |
| Gene length, protein length | 822 bp, 274 aa |
| Immediate neighbours | ndoA, rsbS |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 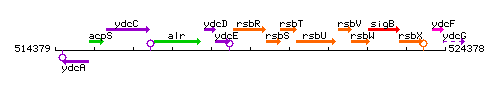
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed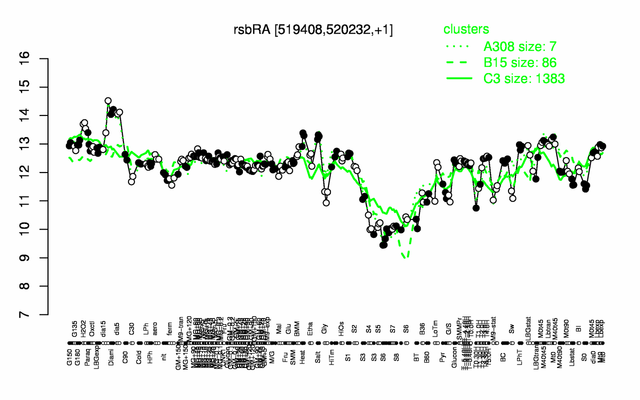
| |
Contents
Categories containing this gene/protein
sigma factors and their control, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU04670
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
Extended information on the protein
- Kinetic information:
- Domains: RsbRA is composed of an N-terminal nonheme globin domain and a highly conserved C-terminal STAS (Sulphate Transporter and AntiSigma factor antagonist) domain. The C-terminal STAS domain is the target of the serine/threonine-specific kinase RsbT (see below).
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P42409
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation: constitutively expressed PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Bill Haldenwang, San Antonio, USA
- Chet Price, Davis, USA homepage
- Rick Lewis, Newcastle, UK homepage
Your additional remarks
References
Reviews
Original Articles
Additional publications: PubMed