Difference between revisions of "CarA"
m (Reverted edits by 134.76.70.252 (talk) to last revision by Jstuelk) |
|||
| Line 102: | Line 102: | ||
* '''Operon:''' ''[[argC]]-[[argJ]]-[[argB]]-[[argD]]-[[carA]]-[[carB]]-[[argF]]'' {{PubMed|7511775}} | * '''Operon:''' ''[[argC]]-[[argJ]]-[[argB]]-[[argD]]-[[carA]]-[[carB]]-[[argF]]'' {{PubMed|7511775}} | ||
| − | * '''[ | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=carA_1199327_1200388_1 carA] {{PubMed|22383849}} |
| + | |||
| + | * '''Sigma factor:''' [[SigA]] {{PubMed|7511775}} | ||
* '''Regulation:''' | * '''Regulation:''' | ||
Revision as of 17:39, 12 April 2012
- Description: carbamoyl-phosphate transferase-arginine (subunit A)
| Gene name | carA |
| Synonyms | cpa |
| Essential | no |
| Product | carbamoyl-phosphate transferase-arginine (subunit A) |
| Function | biosynthesis of arginine |
| Interactions involving this protein in SubtInteract: CarA | |
| Metabolic function and regulation of this protein in SubtiPathways: Pyrimidines, Ammonium/ glutamate | |
| MW, pI | 38 kDa, 6.212 |
| Gene length, protein length | 1059 bp, 353 aa |
| Immediate neighbours | argD, carB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 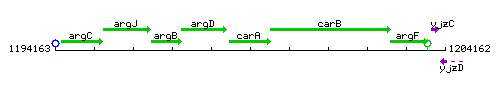
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
biosynthesis/ acquisition of amino acids
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU11230
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
- A mutation was found in this gene after evolution under relaxed selection for sporulation PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2 ATP + L-glutamine + HCO3- + H2O = 2 ADP + phosphate + L-glutamate + carbamoyl phosphate (according to Swiss-Prot)
- Protein family: glutamine amidotransferase type-1 domain (according to Swiss-Prot)
- Paralogous protein(s): PyrAA
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- UniProt: P36838
- KEGG entry: [3]
- E.C. number: 6.3.5.5
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Christopher T Brown, Laura K Fishwick, Binna M Chokshi, Marissa A Cuff, Jay M Jackson, Travis Oglesby, Alison T Rioux, Enrique Rodriguez, Gregory S Stupp, Austin H Trupp, James S Woollcombe-Clarke, Tracy N Wright, William J Zaragoza, Jennifer C Drew, Eric W Triplett, Wayne L Nicholson
Whole-genome sequencing and phenotypic analysis of Bacillus subtilis mutants following evolution under conditions of relaxed selection for sporulation.
Appl Environ Microbiol: 2011, 77(19);6867-77
[PubMed:21821766]
[WorldCat.org]
[DOI]
(I p)
Ulrike Mäder, Georg Homuth, Christian Scharf, Knut Büttner, Rüdiger Bode, Michael Hecker
Transcriptome and proteome analysis of Bacillus subtilis gene expression modulated by amino acid availability.
J Bacteriol: 2002, 184(15);4288-95
[PubMed:12107147]
[WorldCat.org]
[DOI]
(P p)
A Mountain, N H Mann, R N Munton, S Baumberg
Cloning of a Bacillus subtilis restriction fragment complementing auxotrophic mutants of eight Escherichia coli genes of arginine biosynthesis.
Mol Gen Genet: 1984, 197(1);82-9
[PubMed:6096675]
[WorldCat.org]
[DOI]
(P p)