Difference between revisions of "CitG"
| Line 13: | Line 13: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || TCA cycle | |style="background:#ABCDEF;" align="center"|'''Function''' || TCA cycle | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://cellpublisher.gobics.de/subtinteract/startpage/start/ ''Subt''Interact]''': [http://cellpublisher.gobics.de/subtinteract/interactionList/2/CitG CitG] | ||
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/carbon_flow.html Central C-metabolism]''' | ||
Revision as of 17:22, 30 July 2011
- Description: fumarase
| Gene name | citG |
| Synonyms | |
| Essential | no |
| Product | fumarate hydratase |
| Function | TCA cycle |
| Interactions involving this protein in SubtInteract: CitG | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 50 kDa, 5.475 |
| Gene length, protein length | 1386 bp, 462 aa |
| Immediate neighbours | yuxN, gerAA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 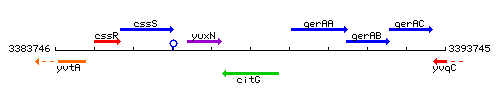
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU33040
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: (S)-malate = fumarate + H2O (according to Swiss-Prot)
- Protein family: Fumarase subfamily (according to Swiss-Prot)
- Paralogous protein(s): AnsB
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure: 1YFE (E.coli)
- UniProt: P07343
- KEGG entry: [3]
- E.C. number: 4.2.1.2
Additional information
Expression and regulation
- Regulation: constitutive
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: GP718 (spec), available in Stülke lab
- Expression vector:
- GFP fusion:
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Stülke lab
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Frederik M Meyer, Jan Gerwig, Elke Hammer, Christina Herzberg, Fabian M Commichau, Uwe Völker, Jörg Stülke
Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon.
Metab Eng: 2011, 13(1);18-27
[PubMed:20933603]
[WorldCat.org]
[DOI]
(I p)
V A Price, I M Feavers, A Moir
Role of sigma H in expression of the fumarase gene (citG) in vegetative cells of Bacillus subtilis 168.
J Bacteriol: 1989, 171(11);5933-9
[PubMed:2509423]
[WorldCat.org]
[DOI]
(P p)
K M Tatti, H L Carter, A Moir, C P Moran
Sigma H-directed transcription of citG in Bacillus subtilis.
J Bacteriol: 1989, 171(11);5928-32
[PubMed:2509422]
[WorldCat.org]
[DOI]
(P p)
I M Feavers, V Price, A Moir
The regulation of the fumarase (citG) gene of Bacillus subtilis 168.
Mol Gen Genet: 1988, 211(3);465-71
[PubMed:3130545]
[WorldCat.org]
[DOI]
(P p)
J S Miles, J R Guest
Complete nucleotide sequence of the fumarase gene (citG) of Bacillus subtilis 168.
Nucleic Acids Res: 1985, 13(1);131-40
[PubMed:3923430]
[WorldCat.org]
[DOI]
(P p)