Difference between revisions of "YosS"
| Line 85: | Line 85: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2BAZ 2BAZ] | + | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=2BAZ 2BAZ] {{PubMed|21358047}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/O34919 O34919] | * '''UniProt:''' [http://www.uniprot.org/uniprot/O34919 O34919] | ||
| Line 127: | Line 127: | ||
=References= | =References= | ||
| − | <pubmed> 19342774 </pubmed> | + | <pubmed> 19342774 21358047 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 15:21, 2 March 2011
- Description: dUTP diphosphatase
| Gene name | yosS |
| Synonyms | |
| Essential | no |
| Product | dUTP diphosphatase |
| Function | nucleotide metabolism |
| MW, pI | 16 kDa, 7.086 |
| Gene length, protein length | 426 bp, 142 aa |
| Immediate neighbours | yosT, yosR |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 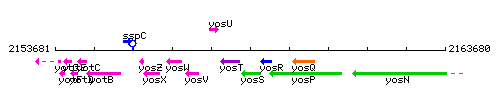
This image was kindly provided by SubtiList
| |
Contents
Categories containing this gene/protein
nucleotide metabolism/ other, SP-beta prophage
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU20020
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): YncF
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- UniProt: O34919
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Javier García-Nafría, Maria Harkiolaki, Rebecca Persson, Mark J Fogg, Keith S Wilson
The structure of Bacillus subtilis SPβ prophage dUTPase and its complexes with two nucleotides.
Acta Crystallogr D Biol Crystallogr: 2011, 67(Pt 3);167-75
[PubMed:21358047]
[WorldCat.org]
[DOI]
(I p)
Gui Lan Li, Juan Wang, Lan Fen Li, Xiao Dong Su
Crystallization and preliminary X-ray analysis of three dUTPases from Gram-positive bacteria.
Acta Crystallogr Sect F Struct Biol Cryst Commun: 2009, 65(Pt 4);339-42
[PubMed:19342774]
[WorldCat.org]
[DOI]
(I p)