Difference between revisions of "YugI"
| Line 1: | Line 1: | ||
| − | * '''Description:''' similar a domain of ''E. coli'' polyribonucleotide phosphorylase and to four repeated domains at the N-terminus of ''E. coli'' ribosomal protein S1 | + | * '''Description:''' similar a domain of ''E. coli'' polyribonucleotide phosphorylase and to four repeated domains at the N-terminus of ''E. coli'' ribosomal protein S1<br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
Revision as of 14:09, 18 May 2010
- Description: similar a domain of E. coli polyribonucleotide phosphorylase and to four repeated domains at the N-terminus of E. coli ribosomal protein S1
| Gene name | yugI |
| Synonyms | |
| Essential | no |
| Product | unknown |
| Function | unknown |
| MW, pI | 14 kDa, 5.918 |
| Gene length, protein length | 390 bp, 130 aa |
| Immediate neighbours | yuzA, alaT |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 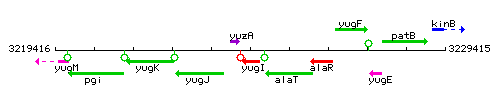
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU31390
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure: 2K4K
- UniProt: P80870
- KEGG entry: [3]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulatory mechanism:
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Wenyu Yu, Bingke Yu, Jicheng Hu, Wei Xia, Changwen Jin, Bin Xia
1H, 13C, and 15N resonance assignments of a general stress protein GSP13 from Bacillus subtilis.
Biomol NMR Assign: 2008, 2(2);163-5
[PubMed:19636895]
[WorldCat.org]
[DOI]
(I p)
Wenyu Yu, Jicheng Hu, Bingke Yu, Wei Xia, Changwen Jin, Bin Xia
Solution structure of GSP13 from Bacillus subtilis exhibits an S1 domain related to cold shock proteins.
J Biomol NMR: 2009, 43(4);255-9
[PubMed:19152054]
[WorldCat.org]
[DOI]
(I p)
Ulf Gerth, Holger Kock, Ilja Kusters, Stephan Michalik, Robert L Switzer, Michael Hecker
Clp-dependent proteolysis down-regulates central metabolic pathways in glucose-starved Bacillus subtilis.
J Bacteriol: 2008, 190(1);321-31
[PubMed:17981983]
[WorldCat.org]
[DOI]
(I p)
Christine Eymann, Georg Homuth, Christian Scharf, Michael Hecker
Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis.
J Bacteriol: 2002, 184(9);2500-20
[PubMed:11948165]
[WorldCat.org]
[DOI]
(P p)
Bauke Oudega, Gregory Koningstein, Luísa Rodrigues, Maria de Sales Ramon, Helmut Hilbert, Andreas Düsterhöft, Thomas M Pohl, Thomas Weitzenegger
Analysis of the Bacillus subtilis genome: cloning and nucleotide sequence of a 62 kb region between 275 degrees (rrnB) and 284 degrees (pai).
Microbiology (Reading): 1997, 143 ( Pt 8);2769-2774
[PubMed:9274030]
[WorldCat.org]
[DOI]
(P p)