Difference between revisions of "Fmt"
(→Database entries) |
|||
| Line 80: | Line 80: | ||
=== Database entries === | === Database entries === | ||
| − | * '''Structure:''' | + | * '''Structure:''' [http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1FMT 1FMT] (from ''Escherichia coli'', 46% identity, 60% similarity) {{PubMed|8887566}} |
* '''UniProt:''' [http://www.uniprot.org/uniprot/P94463 P94463] | * '''UniProt:''' [http://www.uniprot.org/uniprot/P94463 P94463] | ||
Revision as of 16:43, 18 February 2010
- Description: methionyl-tRNA formyltransferase
| Gene name | fmt |
| Synonyms | yloL |
| Essential | yes PubMed |
| Product | methionyl-tRNA formyltransferase |
| Function | formylation of Met-tRNA(fMet) |
| Metabolic function and regulation of this protein in SubtiPathways: tRNA charging | |
| MW, pI | 34 kDa, 5.618 |
| Gene length, protein length | 951 bp, 317 aa |
| Immediate neighbours | def, yloM |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 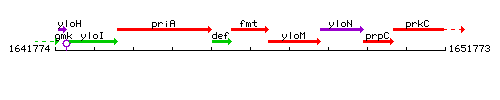
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU15730
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 10-formyltetrahydrofolate + L-methionyl-tRNA(fMet) + H2O = tetrahydrofolate + N-formylmethionyl-tRNA(fMet) (according to Swiss-Prot)
- Protein family: fmt family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- UniProt: P94463
- KEGG entry: [2]
- E.C. number: 2.1.2.9
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Yann Duroc, Carmela Giglione, Thierry Meinnel
Mutations in three distinct loci cause resistance to peptide deformylase inhibitors in Bacillus subtilis.
Antimicrob Agents Chemother: 2009, 53(4);1673-8
[PubMed:19171795]
[WorldCat.org]
[DOI]
(I p)
Krzysztof Hinc, Adam Iwanicki, Simone Seror, Michał Obuchowski
Mapping of a transcription promoter located inside the priA gene of the Bacillus subtilis chromosome.
Acta Biochim Pol: 2006, 53(3);497-505
[PubMed:16964327]
[WorldCat.org]
(P p)