Difference between revisions of "LevD"
| Line 54: | Line 54: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine | + | * '''Catalyzed reaction/ biological activity:''' Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine (according to Swiss-Prot) |
* '''Protein family:''' [[PTS]] permease, mannose permease (Man) family [http://www.ncbi.nlm.nih.gov/pubmed/10627040 PubMed] | * '''Protein family:''' [[PTS]] permease, mannose permease (Man) family [http://www.ncbi.nlm.nih.gov/pubmed/10627040 PubMed] | ||
| Line 89: | Line 89: | ||
=Expression and regulation= | =Expression and regulation= | ||
| + | * '''Operon:''' ''[[levD]]-[[levE]]-[[levF]]-[[levG]]-[[sacC]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/2117666 PubMed] | ||
| − | * ''' | + | * '''[[Sigma factor]]:''' [[SigL]] {{PubMed|1924373}} |
| − | * '''[[ | + | * '''Regulation:''' |
| + | ** subject to carbon catabolite repression ([[CcpA]], [[LevR]]) {{PubMed|7592486}} | ||
| + | ** induced in the presence of fructose ([[LevR]]) {{PubMed|1900939}} | ||
| − | * ''' | + | * '''Regulatory mechanism:''' |
| − | + | ** [[CcpA]]: transcription repression {{PubMed|7592486}} | |
| − | * | + | ** [[LevR]]: transcription activation {{PubMed|1900939}} |
| − | |||
| − | |||
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 122: | Line 123: | ||
=References= | =References= | ||
| − | <pubmed>10627040 2117666 1619665 7592487 7592486 9033408, </pubmed> | + | <pubmed>10627040 2117666 1619665 7592487 7592486 9033408, 1924373 7592486 1900939</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 20:48, 25 November 2009
- Description: fructose-specific phosphotransferase system, EIIA component of the PTS
| Gene name | levD |
| Synonyms | |
| Essential | no |
| Product | fructose-specific phosphotransferase system, EIIA component |
| Function | fructose uptake and phosphorylation |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 16 kDa, 4.479 |
| Gene length, protein length | 438 bp, 146 aa |
| Immediate neighbours | levE, levR |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 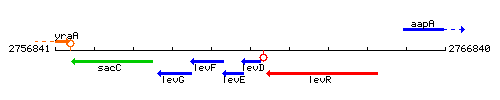
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU27070
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure:
- UniProt: P26379
- KEGG entry: [2]
- E.C. number: 2.7.1.69
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Jonathan Reizer, Steffi Bachem, Aiala Reizer, Maryvonne Arnaud, Milton H Saier, Jörg Stülke
Novel phosphotransferase system genes revealed by genome analysis - the complete complement of PTS proteins encoded within the genome of Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 12);3419-3429
[PubMed:10627040]
[WorldCat.org]
[DOI]
(P p)
V Charrier, J Deutscher, A Galinier, I Martin-Verstraete
Protein phosphorylation chain of a Bacillus subtilis fructose-specific phosphotransferase system and its participation in regulation of the expression of the lev operon.
Biochemistry: 1997, 36(5);1163-72
[PubMed:9033408]
[WorldCat.org]
[DOI]
(P p)
J Stülke, I Martin-Verstraete, V Charrier, A Klier, J Deutscher, G Rapoport
The HPr protein of the phosphotransferase system links induction and catabolite repression of the Bacillus subtilis levanase operon.
J Bacteriol: 1995, 177(23);6928-36
[PubMed:7592487]
[WorldCat.org]
[DOI]
(P p)
I Martin-Verstraete, J Stülke, A Klier, G Rapoport
Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon.
J Bacteriol: 1995, 177(23);6919-27
[PubMed:7592486]
[WorldCat.org]
[DOI]
(P p)
I Martin-Verstraete, M Débarbouillé, A Klier, G Rapoport
Mutagenesis of the Bacillus subtilis "-12, -24" promoter of the levanase operon and evidence for the existence of an upstream activating sequence.
J Mol Biol: 1992, 226(1);85-99
[PubMed:1619665]
[WorldCat.org]
[DOI]
(P p)
M Débarbouillé, I Martin-Verstraete, F Kunst, G Rapoport
The Bacillus subtilis sigL gene encodes an equivalent of sigma 54 from gram-negative bacteria.
Proc Natl Acad Sci U S A: 1991, 88(20);9092-6
[PubMed:1924373]
[WorldCat.org]
[DOI]
(P p)
M Débarbouillé, I Martin-Verstraete, A Klier, G Rapoport
The transcriptional regulator LevR of Bacillus subtilis has domains homologous to both sigma 54- and phosphotransferase system-dependent regulators.
Proc Natl Acad Sci U S A: 1991, 88(6);2212-6
[PubMed:1900939]
[WorldCat.org]
[DOI]
(P p)
I Martin-Verstraete, M Débarbouillé, A Klier, G Rapoport
Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon.
J Mol Biol: 1990, 214(3);657-71
[PubMed:2117666]
[WorldCat.org]
[DOI]
(P p)