Difference between revisions of "SpoIVCB"
(→Extended information on the protein) |
|||
| Line 1: | Line 1: | ||
| − | * '''Description:''' sporulation-specific [[sigma factor]] (SigK) (N-terminal half), other part: ''[[spoIIIC]]'' | + | * '''Description:''' sporulation-specific [[sigma factor]] ([[SigK]]) (N-terminal half), other part: ''[[spoIIIC]]'' |
<br/><br/> | <br/><br/> | ||
| − | |||
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
| Line 96: | Line 95: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' | + | * '''Operon:''' ''[[sigK]]'' (composed of ''[[spoIVCB]]-[[spoIIIC]]'') {{PubMed|2536191}} |
| − | * '''[[Sigma factor]]:''' [[SigE]] {{PubMed|15699190}} | + | * '''[[Sigma factor]]:''' [[SigE]] {{PubMed|15699190,2841290}}, [[SigK]] {{PubMed|2841290}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** expressed during sporulation in the mother cell ([[SigE]], [[SigK]], [[GerE]], [[SpoIIID]]) {{PubMed|15699190,2841290,10075739,7966271}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[GerE]]: transcription repression {{PubMed|10075739}} | ||
| + | ** [[SpoIIID]]: transcription activation {{PubMed|7966271}} | ||
| − | * '''Additional information:''' SigK is activated by processing of membrane-bound pro-SigK by the [[SpoIVFB]] metalloprotease {{PubMed|19805276}} | + | * '''Additional information:''' |
| + | ** SigK is activated by processing of membrane-bound pro-SigK by the [[SpoIVFB]] metalloprotease {{PubMed|19805276}} | ||
| + | ** ''spoIVCB'' and ''[[spoIIIC]]'' form a composite gene in the mother cell during sporulation by excising the intervening DNA sequence. | ||
=Biological materials = | =Biological materials = | ||
| Line 129: | Line 133: | ||
=References= | =References= | ||
| − | <pubmed>14526016,17720779,1900494,10438769,2492118,16385044,1691789,15383836,2536191,2163341,10931284,1577688,9852018,1942049,7814326,8550479,11959848,9501233,10400595,2115401,7592342,1518043,8226681,9078383,2514119,10611287,14523132, 19805276 </pubmed> | + | <pubmed>14526016,17720779,1900494,10438769,2492118,16385044,1691789,15383836,2536191,2163341,10931284,1577688,9852018,1942049,7814326,8550479,11959848,9501233,10400595,2115401,7592342,1518043,8226681,9078383,2514119,10611287,14523132, 19805276 10075739 7966271 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:57, 24 November 2009
- Description: sporulation-specific sigma factor (SigK) (N-terminal half), other part: spoIIIC
| Gene name | spoIVCB |
| Synonyms | cisB, sigK |
| Essential | no |
| Product | RNA polymerase sporulation-specific sigma factor (sigma-K) (N-terminal half) |
| Function | late mother cell-specific gene expression |
| MW, pI | 17 kDa, 8.208 |
| Gene length, protein length | 468 bp, 156 aa |
| Immediate neighbours | nucB, spoIVCA |
| Gene sequence (+200bp) | Protein sequence |
| Caution: The sequence for this gene in SubtiList contains errors | |
Genetic context 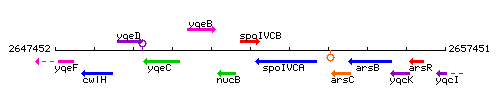
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU25760
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: sigma-70 factor family (according to Swiss-Prot)
- Paralogous protein(s):
Genes controlled by SigK
gerE, lipC, surC, yabG, yndL, yitB-yitA-yisZ
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure:
- UniProt: P12254
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Arthur Aronson, Purdue University, West Lafayette, USA homepage
- Charles Moran, Emory University, NC, USA homepage
Your additional remarks
References
Ruanbao Zhou, Christina Cusumano, Dexin Sui, R Michael Garavito, Lee Kroos
Intramembrane proteolytic cleavage of a membrane-tethered transcription factor by a metalloprotease depends on ATP.
Proc Natl Acad Sci U S A: 2009, 106(38);16174-9
[PubMed:19805276]
[WorldCat.org]
[DOI]
(I p)
Caitlin C Ferguson, Amy H Camp, Richard Losick
gerT, a newly discovered germination gene under the control of the sporulation transcription factor sigmaK in Bacillus subtilis.
J Bacteriol: 2007, 189(21);7681-9
[PubMed:17720779]
[WorldCat.org]
[DOI]
(P p)
Jessica M Silvaggi, John B Perkins, Richard Losick
Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis.
J Bacteriol: 2006, 188(2);532-41
[PubMed:16385044]
[WorldCat.org]
[DOI]
(P p)
Patrick Eichenberger, Masaya Fujita, Shane T Jensen, Erin M Conlon, David Z Rudner, Stephanie T Wang, Caitlin Ferguson, Koki Haga, Tsutomu Sato, Jun S Liu, Richard Losick
The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis.
PLoS Biol: 2004, 2(10);e328
[PubMed:15383836]
[WorldCat.org]
[DOI]
(I p)
Qi Pan, Richard Losick, David Z Rudner
A second PDZ-containing serine protease contributes to activation of the sporulation transcription factor sigmaK in Bacillus subtilis.
J Bacteriol: 2003, 185(20);6051-6
[PubMed:14526016]
[WorldCat.org]
[DOI]
(P p)
Ritsuko Kuwana, Satoko Yamamura, Hiromi Ikejiri, Kazuo Kobayashi, Naotake Ogasawara, Kei Asai, Yoshito Sadaie, Hiromu Takamatsu, Kazuhito Watabe
Bacillus subtilis spoVIF (yjcC) gene, involved in coat assembly and spore resistance.
Microbiology (Reading): 2003, 149(Pt 10);3011-3021
[PubMed:14523132]
[WorldCat.org]
[DOI]
(P p)
David Z Rudner, Richard Losick
A sporulation membrane protein tethers the pro-sigmaK processing enzyme to its inhibitor and dictates its subcellular localization.
Genes Dev: 2002, 16(8);1007-18
[PubMed:11959848]
[WorldCat.org]
[DOI]
(P p)
P R Wakeley, R Dorazi, N T Hoa, J R Bowyer, S M Cutting
Proteolysis of SpolVB is a critical determinant in signalling of Pro-sigmaK processing in Bacillus subtilis.
Mol Microbiol: 2000, 36(6);1336-48
[PubMed:10931284]
[WorldCat.org]
[DOI]
(P p)
D Z Rudner, P Fawcett, R Losick
A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors.
Proc Natl Acad Sci U S A: 1999, 96(26);14765-70
[PubMed:10611287]
[WorldCat.org]
[DOI]
(P p)
J Ju, T Mitchell, H Peters, W G Haldenwang
Sigma factor displacement from RNA polymerase during Bacillus subtilis sporulation.
J Bacteriol: 1999, 181(16);4969-77
[PubMed:10438769]
[WorldCat.org]
[DOI]
(P p)
K H Wade, G Schyns, J A Opdyke, C P Moran
A region of sigmaK involved in promoter activation by GerE in Bacillus subtilis.
J Bacteriol: 1999, 181(14);4365-73
[PubMed:10400595]
[WorldCat.org]
[DOI]
(P p)
H Ichikawa, R Halberg, L Kroos
Negative regulation by the Bacillus subtilis GerE protein.
J Biol Chem: 1999, 274(12);8322-7
[PubMed:10075739]
[WorldCat.org]
[DOI]
(P p)
I Bagyan, B Setlow, P Setlow
New small, acid-soluble proteins unique to spores of Bacillus subtilis: identification of the coding genes and regulation and function of two of these genes.
J Bacteriol: 1998, 180(24);6704-12
[PubMed:9852018]
[WorldCat.org]
[DOI]
(P p)
O Resnekov, R Losick
Negative regulation of the proteolytic activation of a developmental transcription factor in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1998, 95(6);3162-7
[PubMed:9501233]
[WorldCat.org]
[DOI]
(P p)
O Resnekov, S Alper, R Losick
Subcellular localization of proteins governing the proteolytic activation of a developmental transcription factor in Bacillus subtilis.
Genes Cells: 1996, 1(6);529-42
[PubMed:9078383]
[WorldCat.org]
[DOI]
(P p)
H C Carlson, S Lu, L Kroos, W G Haldenwang
Exchange of precursor-specific elements between Pro-sigma E and Pro-sigma K of Bacillus subtilis.
J Bacteriol: 1996, 178(2);546-9
[PubMed:8550479]
[WorldCat.org]
[DOI]
(P p)
C D Webb, A Decatur, A Teleman, R Losick
Use of green fluorescent protein for visualization of cell-specific gene expression and subcellular protein localization during sporulation in Bacillus subtilis.
J Bacteriol: 1995, 177(20);5906-11
[PubMed:7592342]
[WorldCat.org]
[DOI]
(P p)
M Sacco, E Ricca, R Losick, S Cutting
An additional GerE-controlled gene encoding an abundant spore coat protein from Bacillus subtilis.
J Bacteriol: 1995, 177(2);372-7
[PubMed:7814326]
[WorldCat.org]
[DOI]
(P p)
R Halberg, L Kroos
Sporulation regulatory protein SpoIIID from Bacillus subtilis activates and represses transcription by both mother-cell-specific forms of RNA polymerase.
J Mol Biol: 1994, 243(3);425-36
[PubMed:7966271]
[WorldCat.org]
[DOI]
(P p)
V Oke, R Losick
Multilevel regulation of the sporulation transcription factor sigma K in Bacillus subtilis.
J Bacteriol: 1993, 175(22);7341-7
[PubMed:8226681]
[WorldCat.org]
[DOI]
(P p)
L Zheng, R Halberg, S Roels, H Ichikawa, L Kroos, R Losick
Sporulation regulatory protein GerE from Bacillus subtilis binds to and can activate or repress transcription from promoters for mother-cell-specific genes.
J Mol Biol: 1992, 226(4);1037-50
[PubMed:1518043]
[WorldCat.org]
[DOI]
(P p)
E Ricca, S Cutting, R Losick
Characterization of bofA, a gene involved in intercompartmental regulation of pro-sigma K processing during sporulation in Bacillus subtilis.
J Bacteriol: 1992, 174(10);3177-84
[PubMed:1577688]
[WorldCat.org]
[DOI]
(P p)
S Cutting, S Roels, R Losick
Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis.
J Mol Biol: 1991, 221(4);1237-56
[PubMed:1942049]
[WorldCat.org]
[DOI]
(P p)
S Cutting, A Driks, R Schmidt, B Kunkel, R Losick
Forespore-specific transcription of a gene in the signal transduction pathway that governs Pro-sigma K processing in Bacillus subtilis.
Genes Dev: 1991, 5(3);456-66
[PubMed:1900494]
[WorldCat.org]
[DOI]
(P p)
S Cutting, V Oke, A Driks, R Losick, S Lu, L Kroos
A forespore checkpoint for mother cell gene expression during development in B. subtilis.
Cell: 1990, 62(2);239-50
[PubMed:2115401]
[WorldCat.org]
[DOI]
(P p)
L B Zheng, R Losick
Cascade regulation of spore coat gene expression in Bacillus subtilis.
J Mol Biol: 1990, 212(4);645-60
[PubMed:1691789]
[WorldCat.org]
[DOI]
(P p)
B Kunkel, R Losick, P Stragier
The Bacillus subtilis gene for the development transcription factor sigma K is generated by excision of a dispensable DNA element containing a sporulation recombinase gene.
Genes Dev: 1990, 4(4);525-35
[PubMed:2163341]
[WorldCat.org]
[DOI]
(P p)
B Kunkel, L Kroos, H Poth, P Youngman, R Losick
Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis.
Genes Dev: 1989, 3(11);1735-44
[PubMed:2514119]
[WorldCat.org]
[DOI]
(P p)
P Stragier, B Kunkel, L Kroos, R Losick
Chromosomal rearrangement generating a composite gene for a developmental transcription factor.
Science: 1989, 243(4890);507-12
[PubMed:2536191]
[WorldCat.org]
[DOI]
(P p)
L Kroos, B Kunkel, R Losick
Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor.
Science: 1989, 243(4890);526-9
[PubMed:2492118]
[WorldCat.org]
[DOI]
(P p)