Difference between revisions of "GltA"
(→Expression and regulation) |
|||
| Line 99: | Line 99: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[gltA]]-[[gltB]]'' | + | * '''Operon:''' ''[[gltA]]-[[gltB]]'' {{PubMed|12823818}} |
| − | * '''[[Sigma factor]]:''' [[SigA]] | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|2548995}} |
| − | * '''Regulation:''' expression activated by glucose (11 fold) [ | + | * '''Regulation:''' |
| − | ** repressed in the absence of good nitrogen sources (glutamine or ammonium) ([[TnrA]]) | + | ** expression activated by glucose (11 fold) ([[CcpA]], [[GltC]]) {{PubMed|12850135,14523131}} |
| + | ** repressed by arginine ([[GltC]], [[RocG]]) {{PubMed|15150225}} | ||
| + | ** expressed in the presence of ammonium {{PubMed|11029411}} | ||
| + | ** repressed in the absence of good nitrogen sources (glutamine or ammonium) ([[TnrA]]) {{PubMed|11029411}} | ||
| − | * '''Regulatory mechanism:''' | + | * '''Regulatory mechanism:''' |
| − | ** [[TnrA]]: transcription repression | + | ** [[GltC]]: transcription activation {{PubMed|2548995}} |
| + | ** [[TnrA]]: transcription repression {{PubMed|11029411}} | ||
| − | * '''Additional information:''' subject to Clp-dependent proteolysis upon glucose starvation | + | * '''Additional information:''' subject to Clp-dependent proteolysis upon glucose starvation {{PubMed|17981983}} |
=Biological materials = | =Biological materials = | ||
Revision as of 20:29, 18 October 2009
- Description: large subunit of glutamate synthase
| Gene name | gltA |
| Synonyms | |
| Essential | no |
| Product | glutamate synthase (large subunit) |
| Function | glutamate biosynthesis |
| Metabolic function and regulation of this protein in SubtiPathways: Ammonium/ glutamate | |
| MW, pI | 168 kDa, 5.47 |
| Gene length, protein length | 4560 bp, 1520 amino acids |
| Immediate neighbours | gltC, gltB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 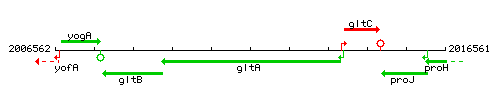
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU18450
Phenotypes of a mutant
auxotrophic for glutamate
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 2 L-glutamate + NADP+ = L-glutamine + 2-oxoglutarate + NADPH (according to Swiss-Prot) 2 L-glutamate + NADP(+) <=> L-glutamine + 2-oxoglutarate + NADPH
- Protein family: glutamate synthase family (according to Swiss-Prot) glutamate synthase family
- Paralogous protein(s): YerD
Extended information on the protein
- Kinetic information:
- Domains:
- Glutamine amidotransferase type-2 domain (22-415)
- Nucleotide binding domain (1060-1112)
- Modification:
- Cofactor(s): 3Fe-4S, FAD, FMN
- Effectors of protein activity:
- Interactions:
- Localization: membrane associated PubMed, cytoplasm
Database entries
- Structure:
- UniProt: P39812
- KEGG entry: [3]
- E.C. number: 1.4.1.13 3 1.4.1.13]
Additional information
subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulation:
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant: GP807 (del gltAB::tet), GP222 (gltA under the control of p-xyl), available in Stülke lab
- Expression vector:
- GFP fusion:
- Antibody:
Labs working on this gene/protein
Linc Sonenshein, Tufts University, Boston, MA, USA Homepage
Jörg Stülke, University of Göttingen, Germany Homepage
Your additional remarks
References