Difference between revisions of "LicH"
| Line 80: | Line 80: | ||
* '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1S6Y 1S6Y] (Geobacillus stearothermophilus) | * '''Structure:''' [http://www.rcsb.org/pdb/explore.do?structureId=1S6Y 1S6Y] (Geobacillus stearothermophilus) | ||
| − | * ''' | + | * '''UniProt:''' [http://www.uniprot.org/uniprot/P46320 P46320] |
* '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU38560] | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU38560] | ||
Revision as of 15:12, 20 July 2009
- Description: 6-phospho-beta-glucosidase
| Gene name | licH |
| Synonyms | celD, celF |
| Essential | no |
| Product | 6-phospho-beta-glucosidase |
| Function | lichenan utilization |
| Metabolic function and regulation of this protein in SubtiPathways: Sugar catabolism | |
| MW, pI | 48 kDa, 5.183 |
| Gene length, protein length | 1326 bp, 442 aa |
| Immediate neighbours | ywaA, licA |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 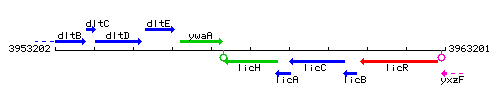
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU38560
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: 6-phospho-beta-D-glucosyl-(1,4)-D-glucose + H2O = D-glucose + D-glucose 6-phosphate (according to Swiss-Prot)
- Protein family: glycosyl hydrolase 4 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure: 1S6Y (Geobacillus stearothermophilus)
- UniProt: P46320
- KEGG entry: [3]
- E.C. number: 3.2.1.86
Additional information
Expression and regulation
- Regulation: repressed by glucose (CcpA) , carbon catabolite repression, induction by oligomeric ß-glucosides PubMed
- Regulatory mechanism: CcpA: transcription repression, catabolite repression: repression by CcpA, induction: transcription activation by the PRD-type regulator LicR PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Takashi Inaoka, Takenori Satomura, Yasutaro Fujita, Kozo Ochi
Novel gene regulation mediated by overproduction of secondary metabolite neotrehalosadiamine in Bacillus subtilis.
FEMS Microbiol Lett: 2009, 291(2);151-6
[PubMed:19087206]
[WorldCat.org]
[DOI]
(I p)
Le Thi Tam, Christine Eymann, Dirk Albrecht, Rabea Sietmann, Frieder Schauer, Michael Hecker, Haike Antelmann
Differential gene expression in response to phenol and catechol reveals different metabolic activities for the degradation of aromatic compounds in Bacillus subtilis.
Environ Microbiol: 2006, 8(8);1408-27
[PubMed:16872404]
[WorldCat.org]
[DOI]
(P p)
S Tobisch, J Stülke, M Hecker
Regulation of the lic operon of Bacillus subtilis and characterization of potential phosphorylation sites of the LicR regulator protein by site-directed mutagenesis.
J Bacteriol: 1999, 181(16);4995-5003
[PubMed:10438772]
[WorldCat.org]
[DOI]
(P p)
S Tobisch, P Glaser, S Krüger, M Hecker
Identification and characterization of a new beta-glucoside utilization system in Bacillus subtilis.
J Bacteriol: 1997, 179(2);496-506
[PubMed:8990303]
[WorldCat.org]
[DOI]
(P p)