Difference between revisions of "ParB"
| Line 86: | Line 86: | ||
* '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P26497 P26497] | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P26497 P26497] | ||
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu:BSU40960] |
* '''E.C. number:''' | * '''E.C. number:''' | ||
Revision as of 04:39, 25 June 2009
- Description: chromosome positioning near the pole and transport through the polar septum / antagonist of Soj-dependent inhibition of sporulation initiation
| Gene name | spo0J |
| Synonyms | spo0JB |
| Essential | no |
| Product | site-specific DNA-binding protein |
| Function | chromosome positioning before asymmetric septation,
specifying its orientation, and imposing directionality on its subsequent transport through the septum; required for sporulation (antagonist of Soj-dependent inhibition of sporulation initiation) |
| MW, pI | 32 kDa, 8.417 |
| Gene length, protein length | 846 bp, 282 aa |
| Immediate neighbours | yyaC, soj |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 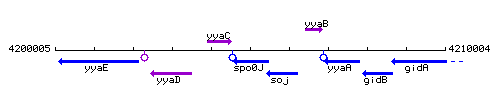
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU40960
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: parB family (according to Swiss-Prot)
- Paralogous protein(s): Noc
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization:
Database entries
- Structure:
- Swiss prot entry: P26497
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Nora L Sullivan, Kathleen A Marquis, David Z Rudner
Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation.
Cell: 2009, 137(4);697-707
[PubMed:19450517]
[WorldCat.org]
[DOI]
(I p)
Stephan Gruber, Jeff Errington
Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis.
Cell: 2009, 137(4);685-96
[PubMed:19450516]
[WorldCat.org]
[DOI]
(I p)
Heath Murray, Jeff Errington
Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA.
Cell: 2008, 135(1);74-84
[PubMed:18854156]
[WorldCat.org]
[DOI]
(I p)
Heath Murray, Henrique Ferreira, Jeff Errington
The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites.
Mol Microbiol: 2006, 61(5);1352-61
[PubMed:16925562]
[WorldCat.org]
[DOI]
(P p)
Gonçalo Real, Sabine Autret, Elizabeth J Harry, Jeffery Errington, Adriano O Henriques
Cell division protein DivIB influences the Spo0J/Soj system of chromosome segregation in Bacillus subtilis.
Mol Microbiol: 2005, 55(2);349-67
[PubMed:15659156]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Ling Juan Wu, Jeff Errington
RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis.
Mol Microbiol: 2003, 49(6);1463-75
[PubMed:12950914]
[WorldCat.org]
[DOI]
(P p)
S Autret, R Nair, J Errington
Genetic analysis of the chromosome segregation protein Spo0J of Bacillus subtilis: evidence for separate domains involved in DNA binding and interactions with Soj protein.
Mol Microbiol: 2001, 41(3);743-55
[PubMed:11532141]
[WorldCat.org]
[DOI]
(P p)
A A Teleman, P L Graumann, D C Lin, A D Grossman, R Losick
Chromosome arrangement within a bacterium.
Curr Biol: 1998, 8(20);1102-9
[PubMed:9778525]
[WorldCat.org]
[DOI]
(P p)
M A Cervin, G B Spiegelman, B Raether, K Ohlsen, M Perego, J A Hoch
A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis.
Mol Microbiol: 1998, 29(1);85-95
[PubMed:9701805]
[WorldCat.org]
[DOI]
(P p)
C D Webb, P L Graumann, J A Kahana, A A Teleman, P A Silver, R Losick
Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis.
Mol Microbiol: 1998, 28(5);883-92
[PubMed:9663676]
[WorldCat.org]
[DOI]
(P p)
P J Lewis, J Errington
Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the SpoOJ partitioning protein.
Mol Microbiol: 1997, 25(5);945-54
[PubMed:9364919]
[WorldCat.org]
[DOI]
(P p)
P Glaser, M E Sharpe, B Raether, M Perego, K Ohlsen, J Errington
Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning.
Genes Dev: 1997, 11(9);1160-8
[PubMed:9159397]
[WorldCat.org]
[DOI]
(P p)
M E Sharpe, J Errington
The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning.
Mol Microbiol: 1996, 21(3);501-9
[PubMed:8866474]
[WorldCat.org]
[DOI]
(P p)
T H Mysliwiec, J Errington, A B Vaidya, M G Bramucci
The Bacillus subtilis spo0J gene: evidence for involvement in catabolite repression of sporulation.
J Bacteriol: 1991, 173(6);1911-9
[PubMed:1900505]
[WorldCat.org]
[DOI]
(P p)