Difference between revisions of "Sandbox"
| Line 1: | Line 1: | ||
| − | * '''Description:''' | + | * '''Description:''' RNA polymerase [[sigma factor]] SigB <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''sigB'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''rpoF '' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || | + | |style="background:#ABCDEF;" align="center"| '''Product''' || RNA polymerase [[sigma factor]] SigB |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Function''' || | + | |style="background:#ABCDEF;" align="center"|'''Function''' || general stress response |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/ | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/pathways/stress_response.html Stress]''' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || | + | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 29 kDa, 5.418 |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || | + | |style="background:#ABCDEF;" align="center"| '''Gene length, protein length''' || 792 bp, 264 aa |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[ | + | |style="background:#ABCDEF;" align="center"|'''Immediate neighbours''' || ''[[rsbW]]'', ''[[rsbX]]'' |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS: | + | |colspan="2" style="background:#FAF8CC;" align="center"|'''Get the DNA and protein [http://srs.ebi.ac.uk/srsbin/cgi-bin/wgetz?-e+[EMBLCDS:CAB12280]+-newId sequences] <br/> (Barbe ''et al.'', 2009)''' |
|- | |- | ||
| − | + | |colspan="2" | '''Genetic context''' <br/> [[Image:sigB_context.gif]] | |
| − | |||
| − | |colspan="2" | '''Genetic context''' <br/> [[Image: | ||
<div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | <div align="right"> <small>This image was kindly provided by [http://genolist.pasteur.fr/SubtiList/ SubtiList]</small></div> | ||
|- | |- | ||
| Line 39: | Line 37: | ||
=== Basic information === | === Basic information === | ||
| − | * '''Locus tag:''' | + | * '''Locus tag:''' BSU04730 |
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| Line 45: | Line 43: | ||
=== Database entries === | === Database entries === | ||
| − | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ | + | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/rsbRSTUVW-sigB-rsbX.html] |
| − | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+ | + | * '''SubtiList entry:''' [http://genolist.pasteur.fr/SubtiList/genome.cgi?gene_detail+BG10735] |
=== Additional information=== | === Additional information=== | ||
| Line 56: | Line 54: | ||
=== Basic information/ Evolution === | === Basic information/ Evolution === | ||
| − | * '''Catalyzed reaction/ biological activity:''' | + | * '''Catalyzed reaction/ biological activity:''' |
| − | * '''Protein family:''' | + | * '''Protein family:''' SigB subfamily (according to Swiss-Prot) |
* '''Paralogous protein(s):''' | * '''Paralogous protein(s):''' | ||
| + | |||
| + | === Genes/ operons controlled by SigB === | ||
| + | |||
| + | ''[[csbB]]'', ''[[ctc]]'', ''[[gsiB]]'', ''[[katE]], [[lexA]],[[licR]], [[sfA]], [[xpf]], [[yaaH]], [[ydeC]], [[ytzE]], [[yvaN]], [[ywhH]]'', | ||
=== Extended information on the protein === | === Extended information on the protein === | ||
| Line 74: | Line 76: | ||
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| − | * '''Interactions:''' [[ | + | * '''Interactions:''' [[SigB]]-[[RsbW]] |
* '''Localization:''' | * '''Localization:''' | ||
| Line 82: | Line 84: | ||
* '''Structure:''' | * '''Structure:''' | ||
| − | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/ | + | * '''Swiss prot entry:''' [http://www.uniprot.org/uniprot/P06574 P06574] |
| − | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+ | + | * '''KEGG entry:''' [http://www.genome.jp/dbget-bin/www_bget?bsu+BSU04730] |
| − | * '''E.C. number:''' | + | * '''E.C. number:''' |
=== Additional information=== | === Additional information=== | ||
| Line 92: | Line 94: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' ''[[ | + | * '''Operon:''' |
| + | ** ''[[rsbR]]-[[rsbS]]-[[rsbT]]-[[rsbU]]-[[rsbV]]-[[rsbW]]-[[sigB]]-[[rsbX]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/8002610 PubMed] | ||
| + | ** ''[[rsbV]]-[[rsbW]]-[[sigB]]-[[rsbX]]'' [http://www.ncbi.nlm.nih.gov/sites/entrez/2170324 PubMed] | ||
| + | |||
| + | * '''[[Sigma factor]]:''' | ||
| + | ** ''[[rsbR]]'': [[SigA]] [http://www.ncbi.nlm.nih.gov/sites/entrez/8002610 PubMed] | ||
| + | ** ''[[rsbV]]:'' [[SigB]] [http://www.ncbi.nlm.nih.gov/pubmed/11544224 PubMed] | ||
| − | * ''' | + | * '''Regulation:''' |
| − | + | ** ''[[rsbV]]:'' induced by stress ([[SigB]]) [http://www.ncbi.nlm.nih.gov/pubmed/11544224 PubMed] | |
| − | |||
| − | |||
| − | |||
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 104: | Line 109: | ||
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' | + | * '''Mutant:''' |
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 117: | Line 122: | ||
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| + | |||
| + | * [[Bill Haldenwang]], San Antonio, USA | ||
| + | * [[Chet Price]], Davis, USA [http://foodscience.ucdavis.edu/price_lab/pricelab.html homepage] | ||
=Your additional remarks= | =Your additional remarks= | ||
| Line 122: | Line 130: | ||
=References= | =References= | ||
| − | <pubmed> | + | <pubmed>8655572,18035607,3123466,8682769,11717291,8460143,10369900,3112122,8468294,8458834,13129942,6784117,12867438,3100810,8253681,12270815,12486038,8764398,3027048,7601843,15342585,17575448,15342582,10482513,8002610,11591687,10220166,17586624,9767581,3016731,11902719,14651641,10503549,15205443,6405278,10383961,15312768,17498739,11377871,15805528 10482513 11544224 11532142 6790515 116131, </pubmed> |
| + | # Höper et al. (2005) Comprehensive Characterization of the Contribution of Individual [[SigB]]-Dependent General Stress Genes to Stress Resistance of ''Bacillus subtilis''. J. Bact. '''187:''' 2810-2826 [http://www.ncbi.nlm.nih.gov/pubmed/15805528 PubMed] | ||
| + | # Petersohn, A., Bernhardt, J., Gerth, U., Höper, D., Koburger, T., Völker, U. and Hecker, M. 1999. Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 181: 5718-5724. [http://www.ncbi.nlm.nih.gov/sites/entrez/10482513 PubMed] | ||
| + | # Petersohn, A., Brigulla, M., Haas, S., Hoheisel, J. D., Völker, U. and Hecker, M. 2001. Global analysis of the general stress reponse of Bacillus subtilis. J. Bacteriol. 183: 5617-5631. [http://www.ncbi.nlm.nih.gov/sites/entrez/11544224 PubMed] | ||
| + | # Price, C. W., Fawcett, P., Cérémonie, H., Su, N., Murphy, C. K. and Youngman, P. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41: 757-774. [http://www.ncbi.nlm.nih.gov/sites/entrez/11532142 PubMed] | ||
| + | # Ollington, J. F., Haldenwang, W. G., Huynh, T. V. and Losick, R. 1981. Developmentally regulated transcription in a cloned segment of the Bacillus subtilis chromosome. J. Bacteriol. 147: 432-442. [http://www.ncbi.nlm.nih.gov/sites/entrez/6790515 PubMed] | ||
| + | # Haldenwang, W. G. & Losick, R. (1979). A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis. Nature 282, 256-260 [http://www.ncbi.nlm.nih.gov/pubmed/116131 PubMed] | ||
# Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | # Author1, Author2 & Author3 (year) Title ''Journal'' '''volume:''' page-page. [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 15:47, 13 June 2009
- Description: RNA polymerase sigma factor SigB
| Gene name | sigB |
| Synonyms | rpoF |
| Essential | no |
| Product | RNA polymerase sigma factor SigB |
| Function | general stress response |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 29 kDa, 5.418 |
| Gene length, protein length | 792 bp, 264 aa |
| Immediate neighbours | rsbW, rsbX |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 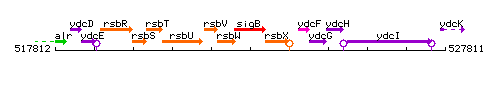
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU04730
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: SigB subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Genes/ operons controlled by SigB
csbB, ctc, gsiB, katE, lexA,licR, sfA, xpf, yaaH, ydeC, ytzE, yvaN, ywhH,
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
Database entries
- Structure:
- Swiss prot entry: P06574
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
- Bill Haldenwang, San Antonio, USA
- Chet Price, Davis, USA homepage
Your additional remarks
References
Michael Hecker, Jan Pané-Farré, Uwe Völker
SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria.
Annu Rev Microbiol: 2007, 61;215-36
[PubMed:18035607]
[WorldCat.org]
[DOI]
(P p)
Adam Reeves, Ulf Gerth, Uwe Völker, W G Haldenwang
ClpP modulates the activity of the Bacillus subtilis stress response transcription factor, sigmaB.
J Bacteriol: 2007, 189(17);6168-75
[PubMed:17586624]
[WorldCat.org]
[DOI]
(P p)
Noriko Suzuki, Naoki Takaya, Takayuki Hoshino, Akira Nakamura
Enhancement of a sigma(B)-dependent stress response in Bacillus subtilis by light via YtvA photoreceptor.
J Gen Appl Microbiol: 2007, 53(2);81-8
[PubMed:17575448]
[WorldCat.org]
[DOI]
(P p)
Oleg A Igoshin, Margaret S Brody, Chester W Price, Michael A Savageau
Distinctive topologies of partner-switching signaling networks correlate with their physiological roles.
J Mol Biol: 2007, 369(5);1333-52
[PubMed:17498739]
[WorldCat.org]
[DOI]
(P p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
Gudrun Holtmann, Matthias Brigulla, Leif Steil, Alexandra Schütz, Karsta Barnekow, Uwe Völker, Erhard Bremer
RsbV-independent induction of the SigB-dependent general stress regulon of Bacillus subtilis during growth at high temperature.
J Bacteriol: 2004, 186(18);6150-8
[PubMed:15342585]
[WorldCat.org]
[DOI]
(P p)
Tae-Jong Kim, Tatiana A Gaidenko, Chester W Price
In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis.
J Bacteriol: 2004, 186(18);6124-32
[PubMed:15342582]
[WorldCat.org]
[DOI]
(P p)
Tae-Jong Kim, Tatiana A Gaidenko, Chester W Price
A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis.
J Mol Biol: 2004, 341(1);135-50
[PubMed:15312768]
[WorldCat.org]
[DOI]
(P p)
Karen Carniol, Tae-Jong Kim, Chester W Price, Richard Losick
Insulation of the sigmaF regulatory system in Bacillus subtilis.
J Bacteriol: 2004, 186(13);4390-4
[PubMed:15205443]
[WorldCat.org]
[DOI]
(P p)
Thorsten Mascher, Neil G Margulis, Tao Wang, Rick W Ye, John D Helmann
Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon.
Mol Microbiol: 2003, 50(5);1591-604
[PubMed:14651641]
[WorldCat.org]
[DOI]
(P p)
Shuyu Zhang, W G Haldenwang
RelA is a component of the nutritional stress activation pathway of the Bacillus subtilis transcription factor sigma B.
J Bacteriol: 2003, 185(19);5714-21
[PubMed:13129942]
[WorldCat.org]
[DOI]
(P p)
Matthias Brigulla, Tamara Hoffmann, Andrea Krisp, Andrea Völker, Erhard Bremer, Uwe Völker
Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation.
J Bacteriol: 2003, 185(15);4305-14
[PubMed:12867438]
[WorldCat.org]
[DOI]
(P p)
Claudia Rollenhagen, Haike Antelmann, Janine Kirstein, Olivier Delumeau, Michael Hecker, Michael D Yudkin
Binding of sigma(A) and sigma(B) to core RNA polymerase after environmental stress in Bacillus subtilis.
J Bacteriol: 2003, 185(1);35-40
[PubMed:12486038]
[WorldCat.org]
[DOI]
(P p)
Olivier Delumeau, Richard J Lewis, Michael D Yudkin
Protein-protein interactions that regulate the energy stress activation of sigma(B) in Bacillus subtilis.
J Bacteriol: 2002, 184(20);5583-9
[PubMed:12270815]
[WorldCat.org]
[DOI]
(P p)
J D Helmann, M F Wu, P A Kobel, F J Gamo, M Wilson, M M Morshedi, M Navre, C Paddon
Global transcriptional response of Bacillus subtilis to heat shock.
J Bacteriol: 2001, 183(24);7318-28
[PubMed:11717291]
[WorldCat.org]
[DOI]
(P p)
M S Brody, K Vijay, C W Price
Catalytic function of an alpha/beta hydrolase is required for energy stress activation of the sigma(B) transcription factor in Bacillus subtilis.
J Bacteriol: 2001, 183(21);6422-8
[PubMed:11591687]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, M Brigulla, S Haas, J D Hoheisel, U Völker, M Hecker
Global analysis of the general stress response of Bacillus subtilis.
J Bacteriol: 2001, 183(19);5617-31
[PubMed:11544224]
[WorldCat.org]
[DOI]
(P p)
C W Price, P Fawcett, H Cérémonie, N Su, C K Murphy, P Youngman
Genome-wide analysis of the general stress response in Bacillus subtilis.
Mol Microbiol: 2001, 41(4);757-74
[PubMed:11532142]
[WorldCat.org]
[DOI]
(P p)
C Eymann, M Hecker
Induction of sigma(B)-dependent general stress genes by amino acid starvation in a spo0H mutant of Bacillus subtilis.
FEMS Microbiol Lett: 2001, 199(2);221-7
[PubMed:11377871]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, S Engelmann, P Setlow, M Hecker
The katX gene of Bacillus subtilis is under dual control of sigmaB and sigmaF.
Mol Gen Genet: 1999, 262(1);173-9
[PubMed:10503549]
[WorldCat.org]
[DOI]
(P p)
A Petersohn, J Bernhardt, U Gerth, D Höper, T Koburger, U Völker, M Hecker
Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization.
J Bacteriol: 1999, 181(18);5718-24
[PubMed:10482513]
[WorldCat.org]
[DOI]
(P p)
U Völker, B Maul, M Hecker
Expression of the sigmaB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis.
J Bacteriol: 1999, 181(13);3942-8
[PubMed:10383961]
[WorldCat.org]
[DOI]
(P p)
T Schweder, A Kolyschkow, U Völker, M Hecker
Analysis of the expression and function of the sigmaB-dependent general stress regulon of Bacillus subtilis during slow growth.
Arch Microbiol: 1999, 171(6);439-43
[PubMed:10369900]
[WorldCat.org]
[DOI]
(P p)
Anja Petersohn, Haike Antelmann, Ulf Gerth, Michael Hecker
Identification and transcriptional analysis of new members of the sigmaB regulon in Bacillus subtilis.
Microbiology (Reading): 1999, 145 ( Pt 4);869-880
[PubMed:10220166]
[WorldCat.org]
[DOI]
(P p)
M Hecker, U Völker
Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon.
Mol Microbiol: 1998, 29(5);1129-36
[PubMed:9767581]
[WorldCat.org]
[DOI]
(P p)
C von Blohn, B Kempf, R M Kappes, E Bremer
Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B.
Mol Microbiol: 1997, 25(1);175-87
[PubMed:11902719]
[WorldCat.org]
[DOI]
(P p)
S Alper, A Dufour, D A Garsin, L Duncan, R Losick
Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis.
J Mol Biol: 1996, 260(2);165-77
[PubMed:8764398]
[WorldCat.org]
[DOI]
(P p)
A Dufour, U Voelker, A Voelker, W G Haldenwang
Relative levels and fractionation properties of Bacillus subtilis σ(B) and its regulators during balanced growth and stress.
J Bacteriol: 1996, 178(13);3701-9 sigma
[PubMed:8682769]
[WorldCat.org]
[DOI]
(P p)
A R Redfield, C W Price
General stress transcription factor sigmaB of Bacillus subtilis is a stable protein.
J Bacteriol: 1996, 178(12);3668-70
[PubMed:8655572]
[WorldCat.org]
[DOI]
(P p)
U Voelker, A Voelker, B Maul, M Hecker, A Dufour, W G Haldenwang
Separate mechanisms activate sigma B of Bacillus subtilis in response to environmental and metabolic stresses.
J Bacteriol: 1995, 177(13);3771-80
[PubMed:7601843]
[WorldCat.org]
[DOI]
(P p)
A A Wise, C W Price
Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor sigma B in response to environmental signals.
J Bacteriol: 1995, 177(1);123-33
[PubMed:8002610]
[WorldCat.org]
[DOI]
(P p)
S A Boylan, A R Redfield, M S Brody, C W Price
Stress-induced activation of the sigma B transcription factor of Bacillus subtilis.
J Bacteriol: 1993, 175(24);7931-7
[PubMed:8253681]
[WorldCat.org]
[DOI]
(P p)
A K Benson, W G Haldenwang
Regulation of sigma B levels and activity in Bacillus subtilis.
J Bacteriol: 1993, 175(8);2347-56
[PubMed:8468294]
[WorldCat.org]
[DOI]
(P p)
A K Benson, W G Haldenwang
The sigma B-dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock.
J Bacteriol: 1993, 175(7);1929-35
[PubMed:8458834]
[WorldCat.org]
[DOI]
(P p)
A K Benson, W G Haldenwang
Bacillus subtilis sigma B is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase.
Proc Natl Acad Sci U S A: 1993, 90(6);2330-4
[PubMed:8460143]
[WorldCat.org]
[DOI]
(P p)
C Ray, M Igo, W Shafer, R Losick, C P Moran
Suppression of ctc promoter mutations in Bacillus subtilis.
J Bacteriol: 1988, 170(2);900-7
[PubMed:3123466]
[WorldCat.org]
[DOI]
(P p)
M Igo, M Lampe, C Ray, W Schafer, C P Moran, R Losick
Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis.
J Bacteriol: 1987, 169(8);3464-9
[PubMed:3112122]
[WorldCat.org]
[DOI]
(P p)
M L Duncan, S S Kalman, S M Thomas, C W Price
Gene encoding the 37,000-dalton minor sigma factor of Bacillus subtilis RNA polymerase: isolation, nucleotide sequence, chromosomal locus, and cryptic function.
J Bacteriol: 1987, 169(2);771-8
[PubMed:3027048]
[WorldCat.org]
[DOI]
(P p)
M M Igo, R Losick
Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis.
J Mol Biol: 1986, 191(4);615-24
[PubMed:3100810]
[WorldCat.org]
[DOI]
(P p)
C Binnie, M Lampe, R Losick
Gene encoding the sigma 37 species of RNA polymerase sigma factor from Bacillus subtilis.
Proc Natl Acad Sci U S A: 1986, 83(16);5943-7
[PubMed:3016731]
[WorldCat.org]
[DOI]
(P p)
W C Johnson, C P Moran, R Losick
Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene.
Nature: 1983, 302(5911);800-4
[PubMed:6405278]
[WorldCat.org]
[DOI]
(P p)
J F Ollington, W G Haldenwang, T V Huynh, R Losick
Developmentally regulated transcription in a cloned segment of the Bacillus subtilis chromosome.
J Bacteriol: 1981, 147(2);432-42
[PubMed:6790515]
[WorldCat.org]
[DOI]
(P p)
W G Haldenwang, R Losick
Novel RNA polymerase sigma factor from Bacillus subtilis.
Proc Natl Acad Sci U S A: 1980, 77(12);7000-4
[PubMed:6784117]
[WorldCat.org]
[DOI]
(P p)
W G Haldenwang, R Losick
A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis.
Nature: 1979, 282(5736);256-60
[PubMed:116131]
[WorldCat.org]
[DOI]
(P p)
- Höper et al. (2005) Comprehensive Characterization of the Contribution of Individual SigB-Dependent General Stress Genes to Stress Resistance of Bacillus subtilis. J. Bact. 187: 2810-2826 PubMed
- Petersohn, A., Bernhardt, J., Gerth, U., Höper, D., Koburger, T., Völker, U. and Hecker, M. 1999. Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 181: 5718-5724. PubMed
- Petersohn, A., Brigulla, M., Haas, S., Hoheisel, J. D., Völker, U. and Hecker, M. 2001. Global analysis of the general stress reponse of Bacillus subtilis. J. Bacteriol. 183: 5617-5631. PubMed
- Price, C. W., Fawcett, P., Cérémonie, H., Su, N., Murphy, C. K. and Youngman, P. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41: 757-774. PubMed
- Ollington, J. F., Haldenwang, W. G., Huynh, T. V. and Losick, R. 1981. Developmentally regulated transcription in a cloned segment of the Bacillus subtilis chromosome. J. Bacteriol. 147: 432-442. PubMed
- Haldenwang, W. G. & Losick, R. (1979). A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis. Nature 282, 256-260 PubMed
- Author1, Author2 & Author3 (year) Title Journal volume: page-page. PubMed