Difference between revisions of "PerR"
(→References) |
|||
| Line 127: | Line 127: | ||
=References= | =References= | ||
| − | <pubmed>11532148, 12029044, 19508285 ,</pubmed> | + | <pubmed> 19508286 ,11532148, 12029044, 19508285 ,</pubmed> |
[http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | [http://www.ncbi.nlm.nih.gov/sites/entrez/PMID PubMed] | ||
Revision as of 12:13, 12 June 2009
- Description: transcriptional repressor of the peroxide regulon
| Gene name | perR |
| Synonyms | ygaG |
| Essential | no |
| Product | transcriptional repressor |
| Function | regulation of the response to peroxide |
| Metabolic function and regulation of this protein in SubtiPathways: Stress | |
| MW, pI | 16 kDa, 5.888 |
| Gene length, protein length | 435 bp, 145 aa |
| Immediate neighbours | ygaF, ygzB |
| Get the DNA and protein sequences (Barbe et al., 2009) | |
Genetic context 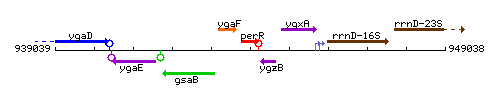
This image was kindly provided by SubtiList
| |
Contents
The gene
Basic information
- Locus tag: BSU08730
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: Fur family (according to Swiss-Prot)
Genes repressed by PerR
Extended information on the protein
- Kinetic information:
- Domains:
- Modification: selective metal catalyzed oxidation of two histidine residues of the regulatory site results in induction (loss of DNA-binding activity) PubMed
- Cofactor(s):
- Effectors of protein activity:
- Interactions:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure: 2RGV
- Swiss prot entry: P71086
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation: negative autoregulation PubMed
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
John Helmann, Cornell University, USA Homepage
Your additional remarks
References
David P Giedroc
Hydrogen peroxide sensing in Bacillus subtilis: it is all about the (metallo)regulator.
Mol Microbiol: 2009, 73(1);1-4
[PubMed:19508286]
[WorldCat.org]
[DOI]
(I p)
L Jacquamet, D A K Traoré, J-L Ferrer, O Proux, D Testemale, J-L Hazemann, E Nazarenko, A El Ghazouani, C Caux-Thang, V Duarte, J-M Latour
Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding.
Mol Microbiol: 2009, 73(1);20-31
[PubMed:19508285]
[WorldCat.org]
[DOI]
(I p)
Mayuree Fuangthong, Andrew F Herbig, Nada Bsat, John D Helmann
Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible.
J Bacteriol: 2002, 184(12);3276-86
[PubMed:12029044]
[WorldCat.org]
[DOI]
(P p)
A F Herbig, J D Helmann
Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA.
Mol Microbiol: 2001, 41(4);849-59
[PubMed:11532148]
[WorldCat.org]
[DOI]
(P p)