Difference between revisions of "MreC"
(→Labs working on this gene/protein) |
Mark Leaver (talk | contribs) |
||
| Line 41: | Line 41: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | ''mreC'' is essential under normal conditions [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed]. Depletion of MreC leads to a progressive increase in the width and a decrease in the length of the cell. This shape defect is consistent with a role for ''mreC'' in cell wall synthesis during elongation and similar to other genes with roles in elongation like [[rodA]] and the redundant gene pair [[pbpA]] and pbpH (also known as [[ykuA]]). Electron microscopy of cells depleted of MreC | + | ''mreC'' is essential under normal conditions [http://www.ncbi.nlm.nih.gov/pubmed/12682299 PubMed]. Depletion of MreC leads to a progressive increase in the width and a decrease in the length of the cell. This shape defect is consistent with a role for ''mreC'' in cell wall synthesis during elongation and has a similar phenotype to other genes with roles in elongation like ''[[rodA]]'' and the redundant gene pair ''[[pbpA]]'' and ''pbpH'' (also known as ''[[ykuA]]''). Electron microscopy of cells depleted of MreC shows regions of the cell where a thick and irregular cell wall has accumulated [http://www.ncbi.nlm.nih.gov/pubmed/12867458 PubMed] and [http://www.ncbi.nlm.nih.gov/pubmed/16101995 PubMed]. ''mreC'' can be deleted provided that 0.5 M sucrose and 20 mM Magnesium is provided in the media, ''mreC'' is therefore conditionally essentail. The phenotype of the ''mreC'' deletion in these conditions is one characterised by extreamly fat and bloated cells that tend to grow in clusters [http://www.ncbi.nlm.nih.gov/pubmed/16101995 PubMed]. |
| + | |||
=== Database entries === | === Database entries === | ||
| Line 54: | Line 55: | ||
===Function=== | ===Function=== | ||
| − | MreC functions in cell wall synthesis by | + | MreC functions in cell wall synthesis by, together with the MreB cytoskeleton, localizing the cell wall synthetic machinery to the correct part of the cell. MreC therefore ensures that the cell wall is made in the correct way to maintain the proper shape of the cell. |
===MreC in other organisms=== | ===MreC in other organisms=== | ||
| − | MreC | + | MreC has been studied in other organisms where it has been shown to be important in cell shape determination. |
| − | *''Escherishia coli'' | + | *''Escherishia coli'' [http://www.ncbi.nlm.nih.gov/pubmed/15612918 PubMed] [http://www.ncbi.nlm.nih.gov/pubmed/17993535 PubMed] |
| − | *''Caulobacter cresentus'' | + | *''Caulobacter cresentus'' [http://www.ncbi.nlm.nih.gov/pubmed/16344480 PubMed] [http://www.ncbi.nlm.nih.gov/pubmed/16344481 PubMed] |
| − | *''Rhodobacter spheroides'' | + | *''Rhodobacter spheroides'' [http://www.ncbi.nlm.nih.gov/pubmed/16484180 PubMed] |
| + | *''Streptomyces coelicolor'' [http://www.ncbi.nlm.nih.gov/pubmed/10954092 PubMed] | ||
| Line 71: | Line 73: | ||
* '''Catalyzed reaction/ biological activity:''' None/ structural protein | * '''Catalyzed reaction/ biological activity:''' None/ structural protein | ||
| − | * '''Protein family:''' | + | * '''Protein family:''' COG1793 |
* '''Paralogous protein(s):''' None | * '''Paralogous protein(s):''' None | ||
| Line 81: | Line 83: | ||
* '''Domains:''' Intracellular N-terminus, transmembrane domain, Coiled coil domain and C-terminal beta-sheet domain. | * '''Domains:''' Intracellular N-terminus, transmembrane domain, Coiled coil domain and C-terminal beta-sheet domain. | ||
| − | * '''Modification:''' | + | * '''Modification:''' None |
| − | * '''Cofactor(s):''' | + | * '''Cofactor(s):''' None |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 137: | Line 139: | ||
=Your additional remarks= | =Your additional remarks= | ||
| + | |||
| + | ''mreC'' is an abbreviation of murein region e, gene C | ||
=References= | =References= | ||
| − | # | + | # Lee JC & Stewart GC (2003) Essential nature of the ''mreC'' determinant of ''Bacillus subtilis'' ''Journal of Bacteriology'' '''185(15)''': 4490-8. [http://www.ncbi.nlm.nih.gov/pubmed/12867458 PubMed] |
| + | # Leaver M & Errington J (2005) Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in ''Bacillus subtilis'' ''Molecular Microbiology'' '''57(5)''': 1196-209 [http://www.ncbi.nlm.nih.gov/pubmed/16101995 PubMed] | ||
| + | # Kruse T, Bork-Jensen J & Gerdes K (2005) The morphogenetic MreBCD proteins of ''Escherichia coli'' form an essential membrane-bound complex ''Molecular Microbiology'' '''55(1)''': 78-89 [http://www.ncbi.nlm.nih.gov/pubmed/15612918 PubMed] | ||
| + | #Bendezú FO & de Boer PA (2008) Conditional lethality, division defects, membrane involution, and endocytosis in ''mre'' and ''mrd'' shape mutants of ''Escherichia coli'' ''Journal of Bacteriology'' '''190(5)''': 1792-811 [http://www.ncbi.nlm.nih.gov/pubmed/17993535 PubMed] | ||
| + | #Divakaruni AV, Loo RR, Xie Y, Loo JA & Gober JW (2005) The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in ''Caulobacter crescentus'' ''PNAS'' '''102(51)''': 18602-7 [http://www.ncbi.nlm.nih.gov/pubmed/16344480 PubMed] | ||
| + | #Dye NA, Pincus Z, Theriot JA, Shapiro L & Gitai Z (2005) Two independent spiral structures control cell shape in Caulobacter ''PNAS'' 102(51): 18608-13. | ||
| + | #Slovak PM, Porter SL & Armitage JP (2006) Differential localization of Mre proteins with PBP2 in ''Rhodobacter sphaeroides'' ''Journal of Bacteriology'' '''188(5)''': 1691-700 | ||
| + | #Burger A, Sichler K, Kelemen G, Buttner M & Wohlleben W (2000) Identification and characterization of the ''mre'' gene region of ''Streptomyces coelicolor'' A3(2). Molecular and General Genetics '''263(6)''': 1053-60 | ||
Revision as of 19:52, 7 March 2009
- Description: MreC is a cell shape determining protein and is associated with the MreB cytoskeleton in B. subtilis and other rod shaped bacteria.
| Gene name | mreC |
| Synonyms | |
| Essential | yes PubMed |
| Product | cell-shape determining protein |
| Function | |
| MW, pI | 32 kDa, 6.248 |
| Gene length, protein length | 870 bp, 290 aa |
| Immediate neighbours | mreD, mreB |
| Gene sequence (+200bp) | Protein sequence |
| Caution: The sequence for this gene in SubtiList contains errors | |
Genetic context 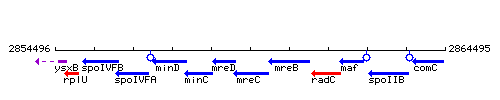
| |
Contents
The gene
Basic information
- Coordinates: 2859062-2859931
Phenotypes of a mutant
mreC is essential under normal conditions PubMed. Depletion of MreC leads to a progressive increase in the width and a decrease in the length of the cell. This shape defect is consistent with a role for mreC in cell wall synthesis during elongation and has a similar phenotype to other genes with roles in elongation like rodA and the redundant gene pair pbpA and pbpH (also known as ykuA). Electron microscopy of cells depleted of MreC shows regions of the cell where a thick and irregular cell wall has accumulated PubMed and PubMed. mreC can be deleted provided that 0.5 M sucrose and 20 mM Magnesium is provided in the media, mreC is therefore conditionally essentail. The phenotype of the mreC deletion in these conditions is one characterised by extreamly fat and bloated cells that tend to grow in clusters PubMed.
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
Function
MreC functions in cell wall synthesis by, together with the MreB cytoskeleton, localizing the cell wall synthetic machinery to the correct part of the cell. MreC therefore ensures that the cell wall is made in the correct way to maintain the proper shape of the cell.
MreC in other organisms
MreC has been studied in other organisms where it has been shown to be important in cell shape determination.
- Escherishia coli PubMed PubMed
- Caulobacter cresentus PubMed PubMed
- Rhodobacter spheroides PubMed
- Streptomyces coelicolor PubMed
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: None/ structural protein
- Protein family: COG1793
- Paralogous protein(s): None
Extended information on the protein
- Kinetic information: None
- Domains: Intracellular N-terminus, transmembrane domain, Coiled coil domain and C-terminal beta-sheet domain.
- Modification: None
- Cofactor(s): None
- Effectors of protein activity:
- Localization: GFP-MreC localises to the cell membrane in a helical pattern PubMed.
Database entries
- Structure:-
- Swiss prot entry: Q01466
- KEGG entry: K03570
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: A non-polar inframe deletion strain named 3481 and a xylose dependent conditional mutant named 3461 is avaliable from the Errington lab PubMed.
- Expression vector:
- lacZ fusion:
- GFP fusion: A functional N-terminal GFP fusion has been made where the fusion protein is the only copy of the gene in the cell: strain 3417 PubMed.
- two-hybrid system:
- Antibody: antisera raised in rabit is avaliable from the Errington lab.
Labs working on this gene/protein
Jeff Errington, Newcastle University, UK homepage
Your additional remarks
mreC is an abbreviation of murein region e, gene C
References
- Lee JC & Stewart GC (2003) Essential nature of the mreC determinant of Bacillus subtilis Journal of Bacteriology 185(15): 4490-8. PubMed
- Leaver M & Errington J (2005) Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis Molecular Microbiology 57(5): 1196-209 PubMed
- Kruse T, Bork-Jensen J & Gerdes K (2005) The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex Molecular Microbiology 55(1): 78-89 PubMed
- Bendezú FO & de Boer PA (2008) Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli Journal of Bacteriology 190(5): 1792-811 PubMed
- Divakaruni AV, Loo RR, Xie Y, Loo JA & Gober JW (2005) The cell-shape protein MreC interacts with extracytoplasmic proteins including cell wall assembly complexes in Caulobacter crescentus PNAS 102(51): 18602-7 PubMed
- Dye NA, Pincus Z, Theriot JA, Shapiro L & Gitai Z (2005) Two independent spiral structures control cell shape in Caulobacter PNAS 102(51): 18608-13.
- Slovak PM, Porter SL & Armitage JP (2006) Differential localization of Mre proteins with PBP2 in Rhodobacter sphaeroides Journal of Bacteriology 188(5): 1691-700
- Burger A, Sichler K, Kelemen G, Buttner M & Wohlleben W (2000) Identification and characterization of the mre gene region of Streptomyces coelicolor A3(2). Molecular and General Genetics 263(6): 1053-60