Difference between revisions of "HemAT"
(→Extended information on the protein) |
|||
| Line 93: | Line 93: | ||
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
** forms clusters at the cell poles {{PubMed|21515776}} | ** forms clusters at the cell poles {{PubMed|21515776}} | ||
| − | ** homogeneous | + | ** homogeneous cytoplasmic distribution in chain-forming cells during the logarithmic phase {{PubMed|23180473}} |
** polar foci in individual cells at later growth stages {{PubMed|23180473}} | ** polar foci in individual cells at later growth stages {{PubMed|23180473}} | ||
Revision as of 14:49, 3 July 2015
- Description: soluble chemotaxis receptor, heme-containing O(2) sensor protein
| Gene name | hemAT |
| Synonyms | yhfV |
| Essential | no |
| Product | haem-based aerotactic transducer |
| Function | movement towards oxygen |
| Gene expression levels in SubtiExpress: hemAT | |
| Interactions involving this protein in SubtInteract: HemAT | |
| MW, pI | 48 kDa, 5.441 |
| Gene length, protein length | 1296 bp, 432 aa |
| Immediate neighbours | yhfU, yhfW |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 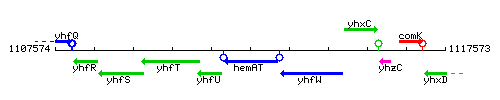
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed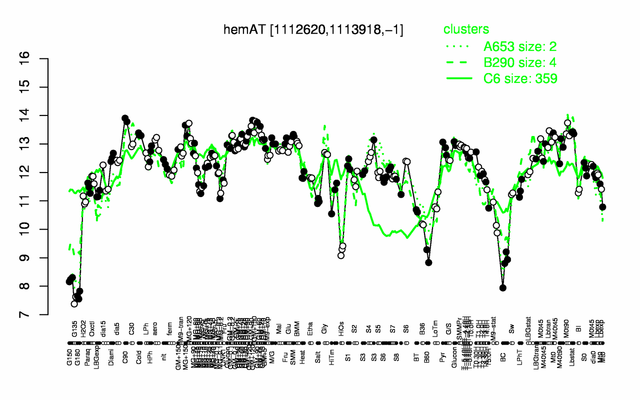
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU10380
Phenotypes of a mutant
- not essential for pellicle biofilm formation, but mutant is outcompeted by the wild-type strain when competed during pellicle formation PubMed
- NCIB3610 hemAT mutant has higher fitness than hag mutant during pellicle formation PubMed
- NCIB3610 hemAT-hag double mutant has similar fitness to single hag mutant during pellicle formation PubMed
Database entries
- BsubCyc: BSU10380
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
- HemAT protein has a moderate oxygen affinity PubMed
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s): heme
- Effectors of protein activity:
Database entries
- BsubCyc: BSU10380
- UniProt: O07621
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: hemAT PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
- in minimal medium, HemAT is present with 19,000 +/- 3,900 molecules per cell PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 1095 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 1803 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 3169 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 966 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 1236 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications
Theresa Hölscher, Benjamin Bartels, Yu-Cheng Lin, Ramses Gallegos-Monterrosa, Alexa Price-Whelan, Roberto Kolter, Lars E P Dietrich, Ákos T Kovács
Motility, Chemotaxis and Aerotaxis Contribute to Competitiveness during Bacterial Pellicle Biofilm Development.
J Mol Biol: 2015, 427(23);3695-3708
[PubMed:26122431]
[WorldCat.org]
[DOI]
(I p)
Carmine G Monteferrante, Calum MacKichan, Elodie Marchadier, Maria-Victoria Prejean, Rut Carballido-López, Jan Maarten van Dijl
Mapping the twin-arginine protein translocation network of Bacillus subtilis.
Proteomics: 2013, 13(5);800-11
[PubMed:23180473]
[WorldCat.org]
[DOI]
(I p)
Yuu Yoshida, Haruto Ishikawa, Shigetoshi Aono, Yasuhisa Mizutani
Structural dynamics of proximal heme pocket in HemAT-Bs associated with oxygen dissociation.
Biochim Biophys Acta: 2012, 1824(7);866-72
[PubMed:22564695]
[WorldCat.org]
[DOI]
(P p)
Vincent J Cannistraro, George D Glekas, Christopher V Rao, George W Ordal
Cellular stoichiometry of the chemotaxis proteins in Bacillus subtilis.
J Bacteriol: 2011, 193(13);3220-7
[PubMed:21515776]
[WorldCat.org]
[DOI]
(I p)
Hideaki Yoshimura, Shiro Yoshioka, Katsuaki Kobayashi, Takehiro Ohta, Takeshi Uchida, Minoru Kubo, Teizo Kitagawa, Shigetoshi Aono
Specific hydrogen-bonding networks responsible for selective O2 sensing of the oxygen sensor protein HemAT from Bacillus subtilis.
Biochemistry: 2006, 45(27);8301-7
[PubMed:16819829]
[WorldCat.org]
[DOI]
(P p)
Masakuni Serizawa, Hiroki Yamamoto, Hirotake Yamaguchi, Yasutaro Fujita, Kazuo Kobayashi, Naotake Ogasawara, Junichi Sekiguchi
Systematic analysis of SigD-regulated genes in Bacillus subtilis by DNA microarray and Northern blotting analyses.
Gene: 2004, 329;125-36
[PubMed:15033535]
[WorldCat.org]
[DOI]
(P p)
S Hou, T Freitas, R W Larsen, M Piatibratov, V Sivozhelezov, A Yamamoto, E A Meleshkevitch, M Zimmer, G W Ordal, M Alam
Globin-coupled sensors: a class of heme-containing sensors in Archaea and Bacteria.
Proc Natl Acad Sci U S A: 2001, 98(16);9353-8
[PubMed:11481493]
[WorldCat.org]
[DOI]
(P p)
S Hou, R W Larsen, D Boudko, C W Riley, E Karatan, M Zimmer, G W Ordal, M Alam
Myoglobin-like aerotaxis transducers in Archaea and Bacteria.
Nature: 2000, 403(6769);540-4
[PubMed:10676961]
[WorldCat.org]
[DOI]
(P p)