Difference between revisions of "KtrC"
(→Biological materials) |
(→Biological materials) |
||
| Line 130: | Line 130: | ||
* '''Mutant:''' | * '''Mutant:''' | ||
** GHB6 (''ktrC''::''spec''), {{PubMed|12562800}} (available in [[Erhard Bremer]]'s and [[Jörg Stülke]]'s) labs, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A955&Search=1A955 BGSC] as 1A955 | ** GHB6 (''ktrC''::''spec''), {{PubMed|12562800}} (available in [[Erhard Bremer]]'s and [[Jörg Stülke]]'s) labs, available at [http://pasture.asc.ohio-state.edu/BGSC/getdetail.cfm?bgscid=1A955&Search=1A955 BGSC] as 1A955 | ||
| + | ** GP2079 (''[[ktrC]]''::''tet''), available in [[Jörg Stülke]]'s lab | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
Revision as of 08:31, 20 July 2015
| Gene name | ktrC |
| Synonyms | ylxV, yzaC, ykqB |
| Essential | no |
| Product | low affinity potassium transporter KtrC-KtrD, peripheric membrane component (proton symport) |
| Function | potassium uptake |
| Gene expression levels in SubtiExpress: ktrC | |
| Interactions involving this protein in SubtInteract: KtrC | |
| Metabolic function and regulation of this protein in SubtiPathways: Metal ion homeostasis, ktrC | |
| MW, pI | 24 kDa, 5.63 |
| Gene length, protein length | 663 bp, 221 aa |
| Immediate neighbours | ykqA, adeC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 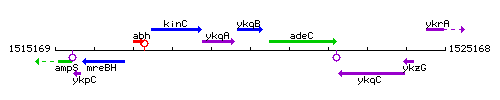
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
transporters/ other, metal ion homeostasis (K, Na, Ca, Mg), coping with hyper-osmotic stress, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU14510
Phenotypes of a mutant
Database entries
- BsubCyc: BSU14510
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): KtrA
Extended information on the protein
- Kinetic information:
- Domains:
- contains a RCK_N domain at the N-terminus (according to UniProt, [2])
- contains a c-di-AMP-binding RCK_C domain at the C-terminus (according to UniProt, [3]) PubMed
- Modification:
- Effectors of protein activity:
- the protein binds c-di-AMP PubMed
- Localization: peripheral membrane protein PubMed
Database entries
- BsubCyc: BSU14510
- Structure:
- UniProt: P39760
- KEGG entry: [4]
- E.C. number:
Additional information
Expression and regulation
- Operon: ktrC PubMed
- Sigma factor:
- Regulation: constitutively expressed PubMed
- Regulatory mechanism:
- Additional information:
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 576 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 453 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 601 PubMed
Biological materials
- Mutant:
- GHB6 (ktrC::spec), PubMed (available in Erhard Bremer's and Jörg Stülke's) labs, available at BGSC as 1A955
- GP2079 (ktrC::tet), available in Jörg Stülke's lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Erhard Bremer, University of Marburg, Germany homepage
Your additional remarks
References
Reviews
Original publications