Difference between revisions of "GlyQ"
(→References) |
(→References) |
||
| Line 148: | Line 148: | ||
=References= | =References= | ||
| − | <pubmed>15890195,15663928,19258532,15292140,16741230,12923262, 15378759 24954903 </pubmed> | + | <pubmed>15890195,15663928,19258532,15292140,16741230,12923262, 15378759 24954903 23589841</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:29, 4 March 2015
- Description: glycyl-tRNA synthetase (alpha subunit)
| Gene name | glyQ |
| Synonyms | yqfJ |
| Essential | yes PubMed |
| Product | glycyl-tRNA synthetase (alpha subunit) |
| Function | translation |
| Gene expression levels in SubtiExpress: glyQ | |
| Interactions involving this protein in SubtInteract: GlyQ | |
| Metabolic function and regulation of this protein in SubtiPathways: glyQ | |
| MW, pI | 33 kDa, 4.8 |
| Gene length, protein length | 885 bp, 295 aa |
| Immediate neighbours | glyS, recO |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 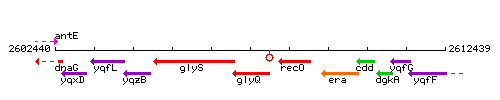
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed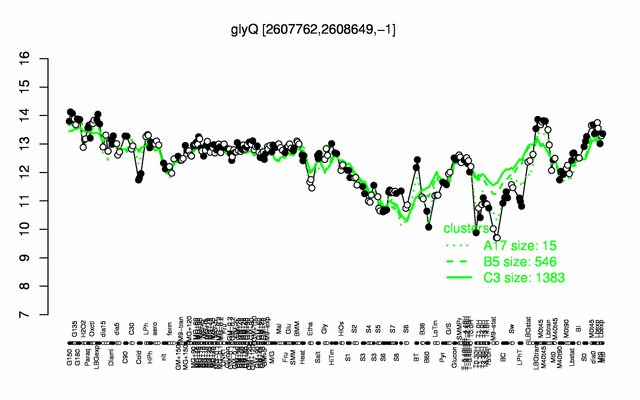
| |
Contents
Categories containing this gene/protein
translation, essential genes, most abundant proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU25270
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU25270
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + glycine + tRNA(Gly) = AMP + diphosphate + glycyl-tRNA(Gly) (according to Swiss-Prot)
- Protein family: class-II aminoacyl-tRNA synthetase family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU25270
- Structure: 1J5W (from Thermotoga maritima, 55% identity, 72% similarity)
- UniProt: P54380
- KEGG entry: [3]
- E.C. number: 6.1.1.14
Additional information
Expression and regulation
- Regulatory mechanism:
- T-box: RNA switch, transcriptional antitermination PubMed
- Additional information:
- belongs to the 100 most abundant proteins PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium): 821 PubMed
- number of protein molecules per cell (complex medium with amino acids, without glucose): 1704 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, exponential phase): 1622 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, early stationary phase after glucose exhaustion): 1214 PubMed
- number of protein molecules per cell (minimal medium with glucose and ammonium, late stationary phase after glucose exhaustion): 1611 PubMed
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References