Difference between revisions of "EzrA"
| Line 129: | Line 129: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
** degradation of [[EzrA]] involves [[FtsH]] {{PubMed|24222488}} | ** degradation of [[EzrA]] involves [[FtsH]] {{PubMed|24222488}} | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 125 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 495 {{PubMed|24696501}} | ||
=Biological materials = | =Biological materials = | ||
Revision as of 09:48, 17 April 2014
- Description: negative regulator of FtsZ ring formation
| Gene name | ezrA |
| Synonyms | ytwP |
| Essential | no |
| Product | FtsZ-interacting protein |
| Function | control of FtsZ ring formation |
| Gene expression levels in SubtiExpress: ezrA | |
| Interactions involving this protein in SubtInteract: EzrA | |
| MW, pI | 64 kDa, 4.757 |
| Gene length, protein length | 1686 bp, 562 aa |
| Immediate neighbours | braB, hisJ |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 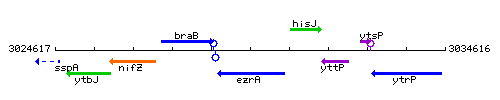
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell division, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU29610
Phenotypes of a mutant
- the ezrA mutation is synthetically lethal with a sepF mutation PubMed
- a yvcL ezrA double mutant grows poorly, and the cells are filamentous PubMed
- a zapA ezrA double mutant forms filamentous cells PubMed
Database entries
- BsubCyc: BSU29610
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: ezrA family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Localization: membrane associated PubMed
- 1 transmembrane domain at the N-terminus according to SOSUI
Database entries
- BsubCyc: BSU29610
- Structure:
- UniProt: O34894
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- constitutively expressed PubMed
- Regulatory mechanism:
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original Publications
Benjamin Mielich-Süss, Johannes Schneider, Daniel Lopez
Overproduction of flotillin influences cell differentiation and shape in Bacillus subtilis.
mBio: 2013, 4(6);e00719-13
[PubMed:24222488]
[WorldCat.org]
[DOI]
(I e)
Ramona Duman, Shu Ishikawa, Ilkay Celik, Henrik Strahl, Naotake Ogasawara, Paulina Troc, Jan Löwe, Leendert W Hamoen
Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring.
Proc Natl Acad Sci U S A: 2013, 110(48);E4601-10
[PubMed:24218584]
[WorldCat.org]
[DOI]
(I p)
Katarina Surdova, Pamela Gamba, Dennis Claessen, Tjalling Siersma, Martijs J Jonker, Jeff Errington, Leendert W Hamoen
The conserved DNA-binding protein WhiA is involved in cell division in Bacillus subtilis.
J Bacteriol: 2013, 195(24);5450-60
[PubMed:24097947]
[WorldCat.org]
[DOI]
(I p)
Erik Nico Trip, Jan-Willem Veening, Eric J Stewart, Jeff Errington, Dirk-Jan Scheffers
Balanced transcription of cell division genes in Bacillus subtilis as revealed by single cell analysis.
Environ Microbiol: 2013, 15(12);3196-209
[PubMed:23701187]
[WorldCat.org]
[DOI]
(I p)
Victoria R Steele, Amy L Bottomley, Jorge Garcia-Lara, Jagath Kasturiarachchi, Simon J Foster
Multiple essential roles for EzrA in cell division of Staphylococcus aureus.
Mol Microbiol: 2011, 80(2);542-55
[PubMed:21401734]
[WorldCat.org]
[DOI]
(I p)
Pamela Gamba, Jan-Willem Veening, Nigel J Saunders, Leendert W Hamoen, Richard A Daniel
Two-step assembly dynamics of the Bacillus subtilis divisome.
J Bacteriol: 2009, 191(13);4186-94
[PubMed:19429628]
[WorldCat.org]
[DOI]
(I p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Dennis Claessen, Robyn Emmins, Leendert W Hamoen, Richard A Daniel, Jeff Errington, David H Edwards
Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis.
Mol Microbiol: 2008, 68(4);1029-46
[PubMed:18363795]
[WorldCat.org]
[DOI]
(I p)
Jay Kumar Singh, Ravindra D Makde, Vinay Kumar, Dulal Panda
A membrane protein, EzrA, regulates assembly dynamics of FtsZ by interacting with the C-terminal tail of FtsZ.
Biochemistry: 2007, 46(38);11013-22
[PubMed:17718511]
[WorldCat.org]
[DOI]
(P p)
Richard B Weart, Amy H Lee, An-Chun Chien, Daniel P Haeusser, Norbert S Hill, Petra Anne Levin
A metabolic sensor governing cell size in bacteria.
Cell: 2007, 130(2);335-47
[PubMed:17662947]
[WorldCat.org]
[DOI]
(P p)
Yoshikazu Kawai, Naotake Ogasawara
Bacillus subtilis EzrA and FtsL synergistically regulate FtsZ ring dynamics during cell division.
Microbiology (Reading): 2006, 152(Pt 4);1129-1141
[PubMed:16549676]
[WorldCat.org]
[DOI]
(P p)
Leendert W Hamoen, Jean-Christophe Meile, Wouter de Jong, Philippe Noirot, Jeff Errington
SepF, a novel FtsZ-interacting protein required for a late step in cell division.
Mol Microbiol: 2006, 59(3);989-99
[PubMed:16420366]
[WorldCat.org]
[DOI]
(P p)
Kuei-Min Chung, Hsin-Hsien Hsu, Suresh Govindan, Ban-Yang Chang
Transcription regulation of ezrA and its effect on cell division of Bacillus subtilis.
J Bacteriol: 2004, 186(17);5926-32
[PubMed:15317798]
[WorldCat.org]
[DOI]
(P p)
David E Anderson, Frederico J Gueiros-Filho, Harold P Erickson
Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins.
J Bacteriol: 2004, 186(17);5775-81
[PubMed:15317782]
[WorldCat.org]
[DOI]
(P p)
Frederico J Gueiros-Filho, Richard Losick
A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ.
Genes Dev: 2002, 16(19);2544-56
[PubMed:12368265]
[WorldCat.org]
[DOI]
(P p)
P A Levin, I G Kurtser, A D Grossman
Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis.
Proc Natl Acad Sci U S A: 1999, 96(17);9642-7
[PubMed:10449747]
[WorldCat.org]
[DOI]
(P p)
Alia Lapidus, Nathalie Galleron, Alexei Sorokin, S Dusko Ehrlich
Sequencing and functional annotation of the Bacillus subtilis genes in the 200 kb rrnB-dnaB region.
Microbiology (Reading): 1997, 143 ( Pt 11);3431-3441
[PubMed:9387221]
[WorldCat.org]
[DOI]
(P p)