Difference between revisions of "PrpC"
| Line 56: | Line 56: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU15760&redirect=T BSU15760] | ||
* '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU15760&redirect=T BSU15760] | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU15760&redirect=T BSU15760] | ||
| Line 95: | Line 96: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU15760&redirect=T BSU15760] | ||
* '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU15760&redirect=T BSU15760] | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU15760&redirect=T BSU15760] | ||
Revision as of 14:10, 2 April 2014
- Description: protein phosphatase
| Gene name | prpC |
| Synonyms | yloO |
| Essential | no |
| Product | protein phosphatase |
| Function | antagonist of PrkC-dependent phosphorylation |
| Gene expression levels in SubtiExpress: prpC | |
| Metabolic function and regulation of this protein in SubtiPathways: prpC | |
| MW, pI | 27 kDa, 4.355 |
| Gene length, protein length | 762 bp, 254 aa |
| Immediate neighbours | yloN, prkC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 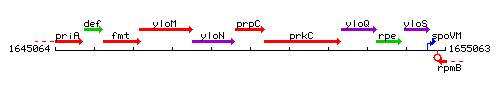
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed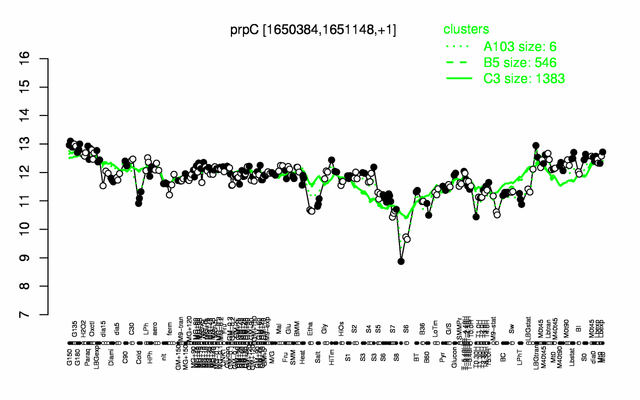
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU15760
Phenotypes of a mutant
A prpC mutant is less lytic in late stationary phase. PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: A phosphoprotein + H2O = a protein + phosphate (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Proteins dephosphorylated by PrpC
CpgA, EF-Tu, YezB PubMed, HPr PubMed, YkwC PubMed
Extended information on the protein
- Kinetic information:
- Modification:
- Cofactors: divalent cations such as magnesium or manganese
- Effectors of protein activity: inhibited by inorganic phosphate and glycero-2-phosphate PubMed
Database entries
- UniProt: O34779
- KEGG entry: [2]
- E.C. number: 3.1.3.16
Additional information
Expression and regulation
- Sigma factor:
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant: OMG401 (aphA3), available in the Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Sandro F F Pereira, Lindsie Goss, Jonathan Dworkin
Eukaryote-like serine/threonine kinases and phosphatases in bacteria.
Microbiol Mol Biol Rev: 2011, 75(1);192-212
[PubMed:21372323]
[WorldCat.org]
[DOI]
(I p)
Original publications
Vaishnavi Ravikumar, Lei Shi, Karsten Krug, Abderahmane Derouiche, Carsten Jers, Charlotte Cousin, Ahasanul Kobir, Ivan Mijakovic, Boris Macek
Quantitative phosphoproteome analysis of Bacillus subtilis reveals novel substrates of the kinase PrkC and phosphatase PrpC.
Mol Cell Proteomics: 2014, 13(8);1965-78
[PubMed:24390483]
[WorldCat.org]
[DOI]
(I p)
Gunjan Arora, Andaleeb Sajid, Mary Diana Arulanandh, Richa Misra, Anshika Singhal, Santosh Kumar, Lalit K Singh, Abid R Mattoo, Rishi Raj, Souvik Maiti, Sharmila Basu-Modak, Yogendra Singh
Zinc regulates the activity of kinase-phosphatase pair (BasPrkC/BasPrpC) in Bacillus anthracis.
Biometals: 2013, 26(5);715-30
[PubMed:23793375]
[WorldCat.org]
[DOI]
(I p)
Cédric Absalon, Michal Obuchowski, Edwige Madec, Delphine Delattre, I Barry Holland, Simone J Séror
CpgA, EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis.
Microbiology (Reading): 2009, 155(Pt 3);932-943
[PubMed:19246764]
[WorldCat.org]
[DOI]
(P p)
Kalpana D Singh, Matthias H Schmalisch, Jörg Stülke, Boris Görke
Carbon catabolite repression in Bacillus subtilis: quantitative analysis of repression exerted by different carbon sources.
J Bacteriol: 2008, 190(21);7275-84
[PubMed:18757537]
[WorldCat.org]
[DOI]
(I p)
Kalpana D Singh, Sven Halbedel, Boris Görke, Jörg Stülke
Control of the phosphorylation state of the HPr protein of the phosphotransferase system in Bacillus subtilis: implication of the protein phosphatase PrpC.
J Mol Microbiol Biotechnol: 2007, 13(1-3);165-71
[PubMed:17693724]
[WorldCat.org]
[DOI]
(P p)
Adam Iwanicki, Krzysztof Hinc, Simone Seror, Grzegorz Wegrzyn, Michal Obuchowski
Transcription in the prpC-yloQ region in Bacillus subtilis.
Arch Microbiol: 2005, 183(6);421-30
[PubMed:16025310]
[WorldCat.org]
[DOI]
(P p)
Kristi E Pullen, Ho-Leung Ng, Pei-Yi Sung, Matthew C Good, Stephen M Smith, Tom Alber
An alternate conformation and a third metal in PstP/Ppp, the M. tuberculosis PP2C-Family Ser/Thr protein phosphatase.
Structure: 2004, 12(11);1947-54
[PubMed:15530359]
[WorldCat.org]
[DOI]
(P p)
Tatiana A Gaidenko, Tae-Jong Kim, Chester W Price
The PrpC serine-threonine phosphatase and PrkC kinase have opposing physiological roles in stationary-phase Bacillus subtilis cells.
J Bacteriol: 2002, 184(22);6109-14
[PubMed:12399479]
[WorldCat.org]
[DOI]
(P p)
M Obuchowski, E Madec, D Delattre, G Boël, A Iwanicki, D Foulger, S J Séror
Characterization of PrpC from Bacillus subtilis, a member of the PPM phosphatase family.
J Bacteriol: 2000, 182(19);5634-8
[PubMed:10986276]
[WorldCat.org]
[DOI]
(P p)