Difference between revisions of "Ldt"
| Line 15: | Line 15: | ||
|- | |- | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU14040 ldt] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU14040 ldt] | ||
| + | |- | ||
| + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=Ldt Ldt]''' | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 17 kDa, 10.252 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 17 kDa, 10.252 | ||
Revision as of 11:43, 8 April 2014
- Description: L,D-transpeptidase involved in cell wall synthesis
| Gene name | ldt |
| Synonyms | ykuD |
| Essential | no |
| Product | L,D-transpeptidase |
| Function | cell wall biosynthesis |
| Gene expression levels in SubtiExpress: ldt | |
| Metabolic function and regulation of this protein in SubtiPathways: Ldt | |
| MW, pI | 17 kDa, 10.252 |
| Gene length, protein length | 492 bp, 164 aa |
| Immediate neighbours | ykuC, ykuE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 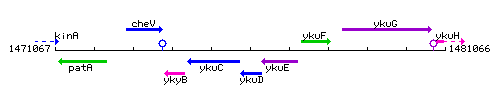
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed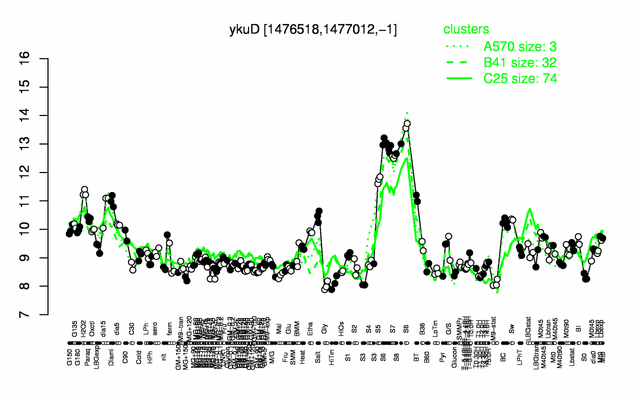
| |
Contents
Categories containing this gene/protein
cell wall synthesis, biosynthesis of cell wall components, sporulation proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU14040
Phenotypes of a mutant
Database entries
- BsubCyc: BSU14040
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- contains a N-acetylglucosamine-polymer-binding LysM domain PubMed
- Modification:
- Effectors of protein activity:
- Localization:
- spore wall (according to Swiss-Prot)
Database entries
- BsubCyc: BSU14040
- Structure: 1Y7M
- UniProt: O34816
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Girbe Buist, Anton Steen, Jan Kok, Oscar P Kuipers
LysM, a widely distributed protein motif for binding to (peptido)glycans.
Mol Microbiol: 2008, 68(4);838-47
[PubMed:18430080]
[WorldCat.org]
[DOI]
(I p)
Original publications
Lauriane Lecoq, Catherine Bougault, Jean-Emmanuel Hugonnet, Carole Veckerlé, Ombeline Pessey, Michel Arthur, Jean-Pierre Simorre
Dynamics induced by β-lactam antibiotics in the active site of Bacillus subtilis L,D-transpeptidase.
Structure: 2012, 20(5);850-61
[PubMed:22579252]
[WorldCat.org]
[DOI]
(I p)
Pierre Nicolas, Ulrike Mäder, Etienne Dervyn, Tatiana Rochat, Aurélie Leduc, Nathalie Pigeonneau, Elena Bidnenko, Elodie Marchadier, Mark Hoebeke, Stéphane Aymerich, Dörte Becher, Paola Bisicchia, Eric Botella, Olivier Delumeau, Geoff Doherty, Emma L Denham, Mark J Fogg, Vincent Fromion, Anne Goelzer, Annette Hansen, Elisabeth Härtig, Colin R Harwood, Georg Homuth, Hanne Jarmer, Matthieu Jules, Edda Klipp, Ludovic Le Chat, François Lecointe, Peter Lewis, Wolfram Liebermeister, Anika March, Ruben A T Mars, Priyanka Nannapaneni, David Noone, Susanne Pohl, Bernd Rinn, Frank Rügheimer, Praveen K Sappa, Franck Samson, Marc Schaffer, Benno Schwikowski, Leif Steil, Jörg Stülke, Thomas Wiegert, Kevin M Devine, Anthony J Wilkinson, Jan Maarten van Dijl, Michael Hecker, Uwe Völker, Philippe Bessières, Philippe Noirot
Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis.
Science: 2012, 335(6072);1103-6
[PubMed:22383849]
[WorldCat.org]
[DOI]
(I p)
L Lecoq, C Bougault, T Kern, J-E Hugonnet, C Veckerlé, O Pessey, M Arthur, J-P Simorre
Backbone and side-chain 1H, 15N and 13C assignment of apo- and imipenem-acylated L,D-transpeptidase from Bacillus subtilis.
Biomol NMR Assign: 2012, 6(2);205-8
[PubMed:22278298]
[WorldCat.org]
[DOI]
(I p)
Sophie Magnet, Ana Arbeloa, Jean-Luc Mainardi, Jean-Emmanuel Hugonnet, Martine Fourgeaud, Lionel Dubost, Arul Marie, Vanessa Delfosse, Claudine Mayer, Louis B Rice, Michel Arthur
Specificity of L,D-transpeptidases from gram-positive bacteria producing different peptidoglycan chemotypes.
J Biol Chem: 2007, 282(18);13151-9
[PubMed:17311917]
[WorldCat.org]
[DOI]
(P p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
T Kodama, H Takamatsu, K Asai, N Ogasawara, Y Sadaie, K Watabe
Synthesis and characterization of the spore proteins of Bacillus subtilis YdhD, YkuD, and YkvP, which carry a motif conserved among cell wall binding proteins.
J Biochem: 2000, 128(4);655-63
[PubMed:11011148]
[WorldCat.org]
[DOI]
(P p)