Difference between revisions of "GcaD"
| Line 128: | Line 128: | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| + | ** number of protein molecules per cell (minimal medium with glucose and ammonium): 397 {{PubMed|24696501}} | ||
| + | ** number of protein molecules per cell (complex medium with amino acids, without glucose): 1922 {{PubMed|24696501}} | ||
=Biological materials = | =Biological materials = | ||
Revision as of 09:51, 17 April 2014
- Description: bifunctional N-acetylglucosamine-1-phosphate uridyltransferase/glucosamine-1-phosphate acetyltransferase
| Gene name | gcaD |
| Synonyms | tms, tms26 |
| Essential | yes PubMed |
| Product | bifunctional N-acetylglucosamine-1-phosphate
uridyltransferase/glucosamine-1-phosphate acetyltransferase |
| Function | cell wall metabolism |
| Gene expression levels in SubtiExpress: gcaD | |
| Metabolic function and regulation of this protein in SubtiPathways: gcaD | |
| MW, pI | 49 kDa, 5.65 |
| Gene length, protein length | 1368 bp, 456 aa |
| Immediate neighbours | spoVG, prs |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 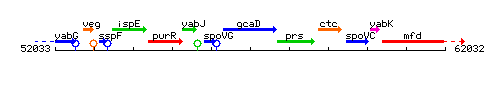
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed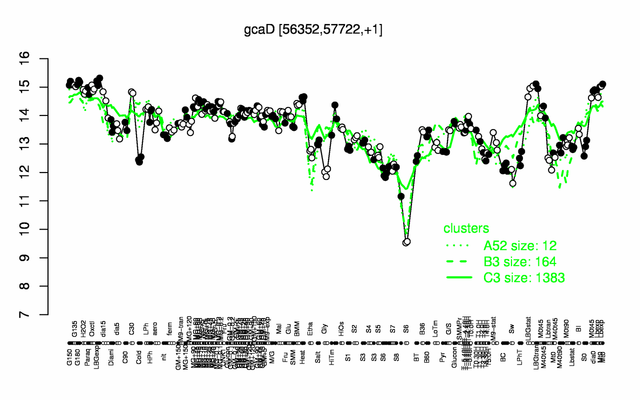
| |
Contents
Categories containing this gene/protein
cell wall synthesis, biosynthesis of cell wall components, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00500
Phenotypes of a mutant
essential PubMed
Database entries
- BsubCyc: BSU00500
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Acetyl-CoA + alpha-D-glucosamine 1-phosphate = CoA + N-acetyl-alpha-D-glucosamine 1-phosphate (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- BsubCyc: BSU00500
- UniProt: P14192
- KEGG entry: [3]
- E.C. number: 2.7.7.23
Additional information
Expression and regulation
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- pGP2596: (IPTG inducible expression, purification in E. coli with N-terminal His-tag, in pWH844), available in Jörg Stülke's lab
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Ludek Sojka, Tomás Kouba, Ivan Barvík, Hana Sanderová, Zdenka Maderová, Jirí Jonák, Libor Krásny
Rapid changes in gene expression: DNA determinants of promoter regulation by the concentration of the transcription initiating NTP in Bacillus subtilis.
Nucleic Acids Res: 2011, 39(11);4598-611
[PubMed:21303765]
[WorldCat.org]
[DOI]
(I p)
G Sulzenbacher, L Gal, C Peneff, F Fassy, Y Bourne
Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture.
J Biol Chem: 2001, 276(15);11844-51
[PubMed:11118459]
[WorldCat.org]
[DOI]
(P p)
I Hilden, B N Krath, B Hove-Jensen
Tricistronic operon expression of the genes gcaD (tms), which encodes N-acetylglucosamine 1-phosphate uridyltransferase, prs, which encodes phosphoribosyl diphosphate synthetase, and ctc in vegetative cells of Bacillus subtilis.
J Bacteriol: 1995, 177(24);7280-4
[PubMed:8522540]
[WorldCat.org]
[DOI]
(P p)
B Hove-Jensen
Identification of tms-26 as an allele of the gcaD gene, which encodes N-acetylglucosamine 1-phosphate uridyltransferase in Bacillus subtilis.
J Bacteriol: 1992, 174(21);6852-6
[PubMed:1328164]
[WorldCat.org]
[DOI]
(P p)