Difference between revisions of "GlxA"
| Line 4: | Line 4: | ||
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Gene name''' | |style="background:#ABCDEF;" align="center"|'''Gene name''' | ||
| − | |'' | + | |''glxA'' |
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''ipa-18r '' | + | |style="background:#ABCDEF;" align="center"| '''Synonyms''' || ''ipa-18r, ywbC '' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
| Line 14: | Line 14: | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || detoxification of methylglyoxal | |style="background:#ABCDEF;" align="center"|'''Function''' || detoxification of methylglyoxal | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU38370 | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Gene expression levels in [http://subtiwiki.uni-goettingen.de/apps/expression/ ''Subti''Express]''': [http://subtiwiki.uni-goettingen.de/apps/expression/expression.php?search=BSU38370 glxA] |
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=ywbC | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=ywbC glxA]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 14 kDa, 4.429 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 14 kDa, 4.429 | ||
| Line 61: | Line 61: | ||
=== Additional information=== | === Additional information=== | ||
| − | |||
=The protein= | =The protein= | ||
| Line 103: | Line 102: | ||
=Expression and regulation= | =Expression and regulation= | ||
| − | * '''Operon:''' '' | + | * '''Operon:''' ''glxA'' {{PubMed|9353933}} |
| − | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ywbC_3937135_3937515_1 | + | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=ywbC_3937135_3937515_1 glxA] {{PubMed|22383849}} |
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
Revision as of 16:44, 14 February 2014
- Description: glyoxalase I
| Gene name | glxA |
| Synonyms | ipa-18r, ywbC |
| Essential | no |
| Product | glyoxalase I |
| Function | detoxification of methylglyoxal |
| Gene expression levels in SubtiExpress: glxA | |
| Metabolic function and regulation of this protein in SubtiPathways: glxA | |
| MW, pI | 14 kDa, 4.429 |
| Gene length, protein length | 378 bp, 126 aa |
| Immediate neighbours | ywbD, ywbB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 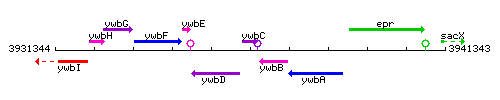
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed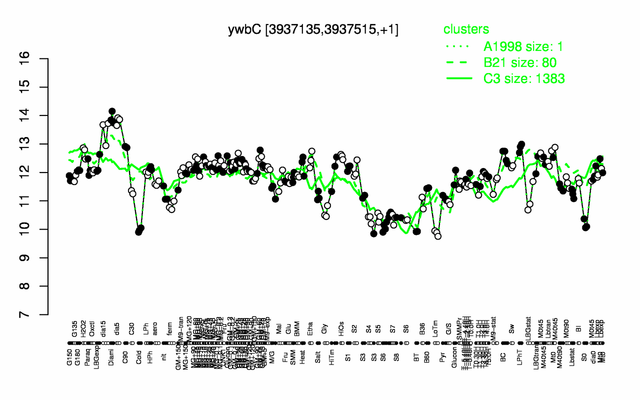
| |
Contents
Categories containing this gene/protein
resistance against oxidative and electrophile stress, poorly characterized/ putative enzymes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU38370
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: glyoxalase I family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P39586
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: glxA PubMed
- Regulation:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Pete Chandrangsu, Renata Dusi, Chris J Hamilton, John D Helmann
Methylglyoxal resistance in Bacillus subtilis: contributions of bacillithiol-dependent and independent pathways.
Mol Microbiol: 2014, 91(4);706-15
[PubMed:24330391]
[WorldCat.org]
[DOI]
(I p)
E Presecan, I Moszer, L Boursier, H Cruz Ramos, V de la Fuente, M-F Hullo, C Lelong, S Schleich, A Sekowska, B H Song, G Villani, F Kunst, A Danchin, P Glaser
The Bacillus subtilis genome from gerBC (311 degrees) to licR (334 degrees).
Microbiology (Reading): 1997, 143 ( Pt 10);3313-3328
[PubMed:9353933]
[WorldCat.org]
[DOI]
(P p)