Difference between revisions of "DivIVA"
(→Original Publications) |
|||
| Line 54: | Line 54: | ||
===Phenotypes of a mutant === | ===Phenotypes of a mutant === | ||
| − | Deletion of divIVA leads to filamentation and polar divisions that in turn cause a minicell phenotype. [http://www.ncbi.nlm.nih.gov/sites/entrez/9219999 PubMed] A divIVA mutant has a severe sporulation defect. [http://www.ncbi.nlm.nih.gov/sites/entrez/11445541 PubMed] | + | * Deletion of ''divIVA'' leads to filamentation and polar divisions that in turn cause a minicell phenotype. [http://www.ncbi.nlm.nih.gov/sites/entrez/9219999 PubMed] |
| + | * A ''divIVA'' mutant has a severe [[sporulation]] defect. [http://www.ncbi.nlm.nih.gov/sites/entrez/11445541 PubMed] | ||
=== Database entries === | === Database entries === | ||
| Line 82: | Line 83: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
** the first 60 amino acids constitute a conserved lipid binding domain. [http://www.ncbi.nlm.nih.gov/sites/entrez/19478798 PubMed] | ** the first 60 amino acids constitute a conserved lipid binding domain. [http://www.ncbi.nlm.nih.gov/sites/entrez/19478798 PubMed] | ||
** the C-terminal domain is less conserved | ** the C-terminal domain is less conserved | ||
| Line 92: | Line 93: | ||
** DivIVA from ''Streptococcus pneumoniae'' is phosphorylated at Threonine 201 by the Ser/Thr protein kinase Sktp1. [http://www.ncbi.nlm.nih.gov/sites/entrez/20453092 PubMed][http://www.ncbi.nlm.nih.gov/sites/entrez/22211696 PubMed] | ** DivIVA from ''Streptococcus pneumoniae'' is phosphorylated at Threonine 201 by the Ser/Thr protein kinase Sktp1. [http://www.ncbi.nlm.nih.gov/sites/entrez/20453092 PubMed][http://www.ncbi.nlm.nih.gov/sites/entrez/22211696 PubMed] | ||
| − | * ''' | + | * '''[[Cofactors]]:''' not known |
* '''Effectors of protein activity:''' not known | * '''Effectors of protein activity:''' not known | ||
| Line 102: | Line 103: | ||
** [[Maf]]-[[DivIVA]] {{PubMed|21564336}} | ** [[Maf]]-[[DivIVA]] {{PubMed|21564336}} | ||
** [[DivIVA]]-[[ComN]] {{PubMed|22582279}} | ** [[DivIVA]]-[[ComN]] {{PubMed|22582279}} | ||
| + | ** [[DivIVA]]-[[SecA]] {{PubMed|24592260}} | ||
* '''[[Localization]]:''' | * '''[[Localization]]:''' | ||
** DivIVA forms a ring underneath the invaginating membrane at the site of cell division and is enriched at both cell poles [http://www.ncbi.nlm.nih.gov/sites/entrez/9219999 PubMed]. | ** DivIVA forms a ring underneath the invaginating membrane at the site of cell division and is enriched at both cell poles [http://www.ncbi.nlm.nih.gov/sites/entrez/9219999 PubMed]. | ||
** forms rings at the division septum and patches at the cell poles {{PubMed|22108385}} | ** forms rings at the division septum and patches at the cell poles {{PubMed|22108385}} | ||
| + | ** membrane targeting requires [[SecA]] {{PubMed|24592260}} | ||
=== Database entries === | === Database entries === | ||
| Line 166: | Line 169: | ||
==Original Publications== | ==Original Publications== | ||
| − | <pubmed>22582279, 19654604, 19666580,9219999,19019154,15554965, 12368265,11445541,10835369,12511520,14651647, 19478798 ,19429628, 11445541, 9219999, 9045828 20352045 20502438 11886553 21564336 22108385 22457634 22517742 22661688 23264578 23701187 23927765 24391905 </pubmed> | + | <pubmed>22582279, 19654604, 19666580,9219999,19019154,15554965, 12368265,11445541,10835369,12511520,14651647, 19478798 ,19429628, 11445541, 9219999, 9045828 20352045 20502438 11886553 21564336 22108385 22457634 22517742 22661688 23264578 23701187 23927765 24391905 24592260 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 08:44, 5 March 2014
- Description: curvature sensitive membrane binding protein that recruits other proteins to the poles and the division septum, cell-division initiation protein (septum placement), part of the Min system (with MinC, MinD, MinJ), Noc and the Min system ensure the efficient utilization of the division site at midcell in by ensuring Z ring placement
| Gene name | divIVA |
| Synonyms | ylmJ |
| Essential | no |
| Product | cell-division initiation protein |
| Function | septum placement |
| Gene expression levels in SubtiExpress: divIVA | |
| Interactions involving this protein in SubtInteract: DivIVA | |
| MW, pI | 19 kDa, 4.846 |
| Gene length, protein length | 492 bp, 164 aa |
| Immediate neighbours | ylmH, ileS |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 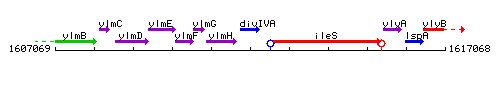
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed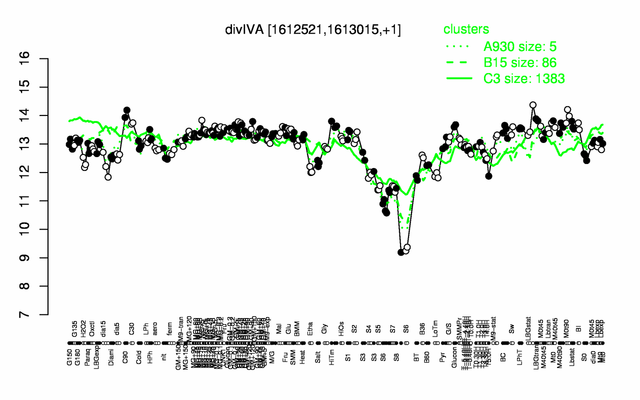
| |
Contents
Categories containing this gene/protein
cell division, membrane proteins, phosphoproteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU15420
Phenotypes of a mutant
- Deletion of divIVA leads to filamentation and polar divisions that in turn cause a minicell phenotype. PubMed
- A divIVA mutant has a severe sporulation defect. PubMed
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
Filamentation is suppressed by mutations in minCD PubMed.
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: gpsB family (according to Swiss-Prot)
- Paralogous protein(s): GpsB
Extended information on the protein
- Kinetic information:
- Modification:
- Cofactors: not known
- Effectors of protein activity: not known
Database entries
- UniProt: P71021
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Operon: divIVA PubMed
- Regulation:
- Additional information:
Biological materials
- Mutant:
- 4041 (divIVA::tet), available in Leendert Hamoen's, Jörg Stülke's, and Sven Halbedel 's lab
- Expression vector: DivIVA-Strep available here
- lacZ fusion:
- GFP fusion: divIVA-gfp fusions available from the Hamoen Lab
- two-hybrid system: B. pertussis adenylate cyclase-based bacterial two hybrid system (BACTH), available in Sven Halbedel's and Jörg Stülke's labs
- Antibody:
Labs working on this gene/protein
Leendert Hamoen, Centre for Bacterial Cell Biology, Newcastle upon Tyne, United Kingdom [4]
Imrich Barak, Slovak Academy of Science, Bratislava, Slovakia homepage
Sven Halbedel, Robert Koch Institute homepage
Your additional remarks
References
Reviews
Original Publications
Sven Halbedel, Maki Kawai, Reinhard Breitling, Leendert W Hamoen
SecA is required for membrane targeting of the cell division protein DivIVA in vivo.
Front Microbiol: 2014, 5;58
[PubMed:24592260]
[WorldCat.org]
[DOI]
(P e)
Lars D Renner, Prahathees Eswaramoorthy, Kumaran S Ramamurthi, Douglas B Weibel
Studying biomolecule localization by engineering bacterial cell wall curvature.
PLoS One: 2013, 8(12);e84143
[PubMed:24391905]
[WorldCat.org]
[DOI]
(I e)
Monika Vishnoi, Jatin Narula, Seram Nganbiton Devi, Hoang-Anh Dao, Oleg A Igoshin, Masaya Fujita
Triggering sporulation in Bacillus subtilis with artificial two-component systems reveals the importance of proper Spo0A activation dynamics.
Mol Microbiol: 2013, 90(1);181-94
[PubMed:23927765]
[WorldCat.org]
[DOI]
(I p)
Erik Nico Trip, Jan-Willem Veening, Eric J Stewart, Jeff Errington, Dirk-Jan Scheffers
Balanced transcription of cell division genes in Bacillus subtilis as revealed by single cell analysis.
Environ Microbiol: 2013, 15(12);3196-209
[PubMed:23701187]
[WorldCat.org]
[DOI]
(I p)
Suey van Baarle, Ilkay Nazli Celik, Karan Gautam Kaval, Marc Bramkamp, Leendert W Hamoen, Sven Halbedel
Protein-protein interaction domains of Bacillus subtilis DivIVA.
J Bacteriol: 2013, 195(5);1012-21
[PubMed:23264578]
[WorldCat.org]
[DOI]
(I p)
Joe Pogliano, Nicolas Pogliano, Jared A Silverman
Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins.
J Bacteriol: 2012, 194(17);4494-504
[PubMed:22661688]
[WorldCat.org]
[DOI]
(I p)
Valquiria Tiago dos Santos, Alexandre W Bisson-Filho, Frederico J Gueiros-Filho
DivIVA-mediated polar localization of ComN, a posttranscriptional regulator of Bacillus subtilis.
J Bacteriol: 2012, 194(14);3661-9
[PubMed:22582279]
[WorldCat.org]
[DOI]
(I p)
Alexander K W Elsholz, Kürsad Turgay, Stephan Michalik, Bernd Hessling, Katrin Gronau, Dan Oertel, Ulrike Mäder, Jörg Bernhardt, Dörte Becher, Michael Hecker, Ulf Gerth
Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis.
Proc Natl Acad Sci U S A: 2012, 109(19);7451-6
[PubMed:22517742]
[WorldCat.org]
[DOI]
(I p)
Christopher D A Rodrigues, Elizabeth J Harry
The Min system and nucleoid occlusion are not required for identifying the division site in Bacillus subtilis but ensure its efficient utilization.
PLoS Genet: 2012, 8(3);e1002561
[PubMed:22457634]
[WorldCat.org]
[DOI]
(I p)
Prahathees Eswaramoorthy, Marcella L Erb, James A Gregory, Jared Silverman, Kit Pogliano, Joe Pogliano, Kumaran S Ramamurthi
Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis.
mBio: 2011, 2(6);
[PubMed:22108385]
[WorldCat.org]
[DOI]
(I e)
Kenneth Briley, Peter Prepiak, Miguel J Dias, Jeanette Hahn, David Dubnau
Maf acts downstream of ComGA to arrest cell division in competent cells of B. subtilis.
Mol Microbiol: 2011, 81(1);23-39
[PubMed:21564336]
[WorldCat.org]
[DOI]
(I p)
Maria A Oliva, Sven Halbedel, Stefan M Freund, Pavel Dutow, Thomas A Leonard, Dmitry B Veprintsev, Leendert W Hamoen, Jan Löwe
Features critical for membrane binding revealed by DivIVA crystal structure.
EMBO J: 2010, 29(12);1988-2001
[PubMed:20502438]
[WorldCat.org]
[DOI]
(I p)
Suey van Baarle, Marc Bramkamp
The MinCDJ system in Bacillus subtilis prevents minicell formation by promoting divisome disassembly.
PLoS One: 2010, 5(3);e9850
[PubMed:20352045]
[WorldCat.org]
[DOI]
(I e)
Kumaran S Ramamurthi, Richard Losick
Negative membrane curvature as a cue for subcellular localization of a bacterial protein.
Proc Natl Acad Sci U S A: 2009, 106(32);13541-5
[PubMed:19666580]
[WorldCat.org]
[DOI]
(I p)
Jennifer R Juarez, William Margolin
Irresistible curves.
EMBO J: 2009, 28(15);2147-8
[PubMed:19654604]
[WorldCat.org]
[DOI]
(I p)
Rok Lenarcic, Sven Halbedel, Loek Visser, Michael Shaw, Ling Juan Wu, Jeff Errington, Davide Marenduzzo, Leendert W Hamoen
Localisation of DivIVA by targeting to negatively curved membranes.
EMBO J: 2009, 28(15);2272-82
[PubMed:19478798]
[WorldCat.org]
[DOI]
(I p)
Pamela Gamba, Jan-Willem Veening, Nigel J Saunders, Leendert W Hamoen, Richard A Daniel
Two-step assembly dynamics of the Bacillus subtilis divisome.
J Bacteriol: 2009, 191(13);4186-94
[PubMed:19429628]
[WorldCat.org]
[DOI]
(I p)
Marc Bramkamp, Robyn Emmins, Louise Weston, Catriona Donovan, Richard A Daniel, Jeff Errington
A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD.
Mol Microbiol: 2008, 70(6);1556-69
[PubMed:19019154]
[WorldCat.org]
[DOI]
(I p)
S E Perry, D H Edwards
Identification of a polar targeting determinant for Bacillus subtilis DivIVA.
Mol Microbiol: 2004, 54(5);1237-49
[PubMed:15554965]
[WorldCat.org]
[DOI]
(P p)
Virginie Molle, Masaya Fujita, Shane T Jensen, Patrick Eichenberger, José E González-Pastor, Jun S Liu, Richard Losick
The Spo0A regulon of Bacillus subtilis.
Mol Microbiol: 2003, 50(5);1683-701
[PubMed:14651647]
[WorldCat.org]
[DOI]
(P p)
Leendert W Hamoen, Jeffery Errington
Polar targeting of DivIVA in Bacillus subtilis is not directly dependent on FtsZ or PBP 2B.
J Bacteriol: 2003, 185(2);693-7
[PubMed:12511520]
[WorldCat.org]
[DOI]
(P p)
Frederico J Gueiros-Filho, Richard Losick
A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ.
Genes Dev: 2002, 16(19);2544-56
[PubMed:12368265]
[WorldCat.org]
[DOI]
(P p)
M E Karoui, J Errington
Isolation and characterization of topological specificity mutants of minD in Bacillus subtilis.
Mol Microbiol: 2001, 42(5);1211-21
[PubMed:11886553]
[WorldCat.org]
[DOI]
(P p)
H B Thomaides, M Freeman, M El Karoui, J Errington
Division site selection protein DivIVA of Bacillus subtilis has a second distinct function in chromosome segregation during sporulation.
Genes Dev: 2001, 15(13);1662-73
[PubMed:11445541]
[WorldCat.org]
[DOI]
(P p)
D H Edwards, H B Thomaides, J Errington
Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast.
EMBO J: 2000, 19(11);2719-27
[PubMed:10835369]
[WorldCat.org]
[DOI]
(P p)
D H Edwards, J Errington
The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division.
Mol Microbiol: 1997, 24(5);905-15
[PubMed:9219999]
[WorldCat.org]
[DOI]
(P p)
J H Cha, G C Stewart
The divIVA minicell locus of Bacillus subtilis.
J Bacteriol: 1997, 179(5);1671-83
[PubMed:9045828]
[WorldCat.org]
[DOI]
(P p)