Difference between revisions of "BglS"
| Line 79: | Line 79: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 127: | Line 127: | ||
=Biological materials = | =Biological materials = | ||
| − | * '''Mutant:''' GP427 (licT-bglS, erm), BGW7 (cat), both available in the [[Stülke]] lab | + | * '''Mutant:''' GP427 (''[[licT]]-[[bglS]]'', erm), BGW7 (cat), both available in the [[Stülke]] lab |
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 145: | Line 145: | ||
=References= | =References= | ||
| − | <pubmed>18957862 12850135 6087283 8245830 8245831 8606172, 23459129 </pubmed> | + | <pubmed>18957862 12850135 6087283 8245830 8245831 8606172, 23459129 24391357 </pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:22, 7 January 2014
- Description: endo-beta-1,3-1,4 glucanase
| Gene name | bglS |
| Synonyms | bgl, licS |
| Essential | no |
| Product | endo-beta-1,3-1,4 glucanase |
| Function | lichenan degradation |
| Gene expression levels in SubtiExpress: bglS | |
| Metabolic function and regulation of this protein in SubtiPathways: bglS | |
| MW, pI | 27 kDa, 6.482 |
| Gene length, protein length | 726 bp, 242 aa |
| Immediate neighbours | citH, licT |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed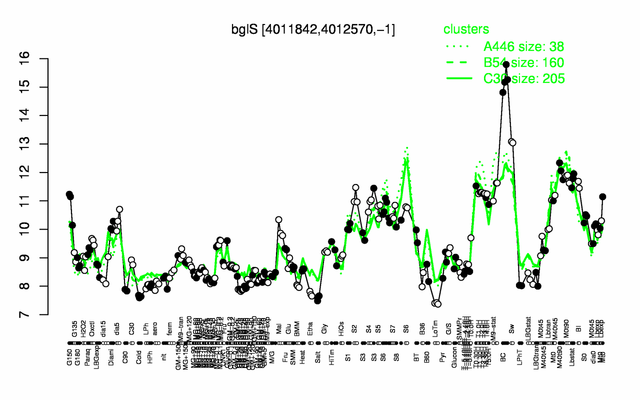
| |
Contents
Categories containing this gene/protein
utilization of specific carbon sources
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU39070
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: Hydrolysis of (1->4)-beta-D-glucosidic linkages in beta-D-glucans containing (1->3)- and (1->4)-bonds (according to Swiss-Prot)
- Protein family: glycosyl hydrolase 16 family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization: extracellular (signal peptide) PubMed
Database entries
- Structure: 3O5S
- UniProt: P04957
- KEGG entry: [3]
- E.C. number: 3.2.1.73
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- CcpA: transcription repression PubMed
- LicT: binding to an RNA switch results in transcriptional antitermination PubMed
- Additional information:
Biological materials
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Revathi Masilamani, Om Prakash Sharma, Suresh Kumar Muthuvel, Sakthivel Natarajan
Cloning, expression of b-1,3-1,4 glucanase from Bacillus subtilis SU40 and the effect of calcium ion on the stability of recombinant enzyme: in vitro and in silico analysis.
Bioinformation: 2013, 9(19);958-62
[PubMed:24391357]
[WorldCat.org]
[DOI]
(P e)
Junio Cota, Leandro C Oliveira, André R L Damásio, Ana P Citadini, Zaira B Hoffmam, Thabata M Alvarez, Carla A Codima, Vitor B P Leite, Glaucia Pastore, Mario de Oliveira-Neto, Mario T Murakami, Roberto Ruller, Fabio M Squina
Assembling a xylanase-lichenase chimera through all-atom molecular dynamics simulations.
Biochim Biophys Acta: 2013, 1834(8);1492-500
[PubMed:23459129]
[WorldCat.org]
[DOI]
(P p)
Birgit Voigt, Haike Antelmann, Dirk Albrecht, Armin Ehrenreich, Karl-Heinz Maurer, Stefan Evers, Gerhard Gottschalk, Jan Maarten van Dijl, Thomas Schweder, Michael Hecker
Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis.
J Mol Microbiol Biotechnol: 2009, 16(1-2);53-68
[PubMed:18957862]
[WorldCat.org]
[DOI]
(I p)
Hans-Matti Blencke, Georg Homuth, Holger Ludwig, Ulrike Mäder, Michael Hecker, Jörg Stülke
Transcriptional profiling of gene expression in response to glucose in Bacillus subtilis: regulation of the central metabolic pathways.
Metab Eng: 2003, 5(2);133-49
[PubMed:12850135]
[WorldCat.org]
[DOI]
(P p)
K Schnetz, J Stülke, S Gertz, S Krüger, M Krieg, M Hecker, B Rak
LicT, a Bacillus subtilis transcriptional antiterminator protein of the BglG family.
J Bacteriol: 1996, 178(7);1971-9
[PubMed:8606172]
[WorldCat.org]
[DOI]
(P p)
S Krüger, J Stülke, M Hecker
Catabolite repression of beta-glucanase synthesis in Bacillus subtilis.
J Gen Microbiol: 1993, 139(9);2047-54
[PubMed:8245831]
[WorldCat.org]
[DOI]
(P p)
J Stülke, R Hanschke, M Hecker
Temporal activation of beta-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool.
J Gen Microbiol: 1993, 139(9);2041-5
[PubMed:8245830]
[WorldCat.org]
[DOI]
(P p)
N Murphy, D J McConnell, B A Cantwell
The DNA sequence of the gene and genetic control sites for the excreted B. subtilis enzyme beta-glucanase.
Nucleic Acids Res: 1984, 12(13);5355-67
[PubMed:6087283]
[WorldCat.org]
[DOI]
(P p)