Difference between revisions of "FtsH"
| Line 64: | Line 64: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU00690&redirect=T BSU00690] | ||
* '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ftsH.html] | * '''DBTBS entry:''' [http://dbtbs.hgc.jp/COG/prom/ftsH.html] | ||
| Line 110: | Line 111: | ||
=== Database entries === | === Database entries === | ||
| + | * '''BsubCyc:''' [http://bsubcyc.org/BSUB/NEW-IMAGE?type=NIL&object=BSU00690&redirect=T BSU00690] | ||
* '''Structure:''' [http://www.pdb.org/pdb/explore/explore.do?structureId=1LV7 1LV7] (from ''E. coli'', 64% identity) {{PubMed|12176385}} | * '''Structure:''' [http://www.pdb.org/pdb/explore/explore.do?structureId=1LV7 1LV7] (from ''E. coli'', 64% identity) {{PubMed|12176385}} | ||
Revision as of 12:47, 2 April 2014
- Description: ATP-dependent metalloprotease
| Gene name | ftsH |
| Synonyms | |
| Essential | no |
| Product | ATP-dependent metalloprotease |
| Function | cell division, sporulation initiation |
| Gene expression levels in SubtiExpress: ftsH | |
| Interactions involving this protein in SubtInteract: FtsH | |
| Metabolic function and regulation of this protein in SubtiPathways: ftsH | |
| MW, pI | 70 kDa, 5.841 |
| Gene length, protein length | 1911 bp, 637 aa |
| Immediate neighbours | hprT, coaX |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 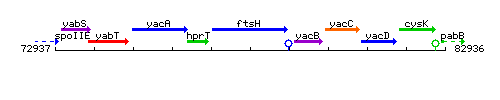
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed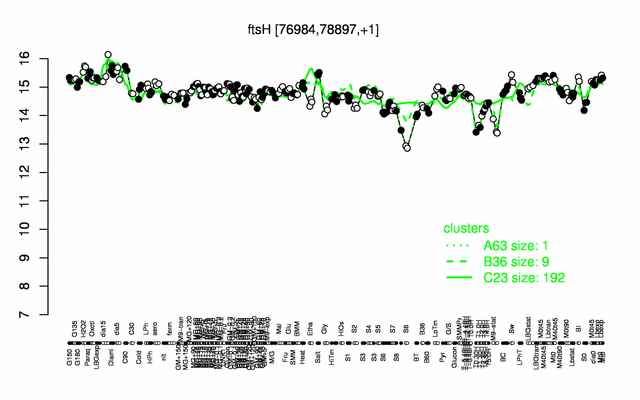
| |
Contents
Categories containing this gene/protein
cell division, proteolysis, cell envelope stress proteins (controlled by SigM, V, W, X, Y), heat shock proteins, biofilm formation, membrane proteins
This gene is a member of the following regulons
SigM regulon, TilS-HprT regulon
The gene
Basic information
- Locus tag: BSU00690
Phenotypes of a mutant
Database entries
- BsubCyc: BSU00690
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): YjoB
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- BsubCyc: BSU00690
- UniProt: P37476
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Regulation: induced by heat shock (class III)
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Noël Molière, Kürşad Turgay
General and regulatory proteolysis in Bacillus subtilis.
Subcell Biochem: 2013, 66;73-103
[PubMed:23479438]
[WorldCat.org]
[DOI]
(P p)
Original publications
Benjamin Mielich-Süss, Johannes Schneider, Daniel Lopez
Overproduction of flotillin influences cell differentiation and shape in Bacillus subtilis.
mBio: 2013, 4(6);e00719-13
[PubMed:24222488]
[WorldCat.org]
[DOI]
(I e)
Ta-Hui Lin, Yi-Nei Hu, Gwo-Chyuan Shaw
Two enzymes, TilS and HprT, can form a complex to function as a transcriptional activator for the cell division protease gene ftsH in Bacillus subtilis.
J Biochem: 2014, 155(1);5-16
[PubMed:24001521]
[WorldCat.org]
[DOI]
(I p)
Ana Yepes, Johannes Schneider, Benjamin Mielich, Gudrun Koch, Juan-Carlos García-Betancur, Kumaran S Ramamurthi, Hera Vlamakis, Daniel López
The biofilm formation defect of a Bacillus subtilis flotillin-defective mutant involves the protease FtsH.
Mol Microbiol: 2012, 86(2);457-71
[PubMed:22882210]
[WorldCat.org]
[DOI]
(I p)
Hue Bach Thi Nguyen, Wolfgang Schumann
The sporulation control gene spo0M of Bacillus subtilis is a target of the FtsH metalloprotease.
Res Microbiol: 2012, 163(2);114-8
[PubMed:22142536]
[WorldCat.org]
[DOI]
(I p)
Ai Thi Thuy Le, Wolfgang Schumann
The Spo0E phosphatase of Bacillus subtilis is a substrate of the FtsH metalloprotease.
Microbiology (Reading): 2009, 155(Pt 4);1122-1132
[PubMed:19332814]
[WorldCat.org]
[DOI]
(P p)
Hannes Hahne, Susanne Wolff, Michael Hecker, Dörte Becher
From complementarity to comprehensiveness--targeting the membrane proteome of growing Bacillus subtilis by divergent approaches.
Proteomics: 2008, 8(19);4123-36
[PubMed:18763711]
[WorldCat.org]
[DOI]
(I p)
Warawan Eiamphungporn, John D Helmann
The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses.
Mol Microbiol: 2008, 67(4);830-48
[PubMed:18179421]
[WorldCat.org]
[DOI]
(P p)
Matthias Kotschwar, Evita Harfst, Tanja Ohanjan, Wolfgang Schumann
Construction and analyses of mutant ftsH alleles of Bacillus subtilis involving the ATPase- and Zn-binding domains.
Curr Microbiol: 2004, 49(3);180-5
[PubMed:15386101]
[WorldCat.org]
[DOI]
(P p)
G Hambraeus, C von Wachenfeldt, L Hederstedt
Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs.
Mol Genet Genomics: 2003, 269(5);706-14
[PubMed:12884008]
[WorldCat.org]
[DOI]
(P p)
Stephan Zellmeier, Ulrich Zuber, Wolfgang Schumann, Thomas Wiegert
The absence of FtsH metalloprotease activity causes overexpression of the sigmaW-controlled pbpE gene, resulting in filamentous growth of Bacillus subtilis.
J Bacteriol: 2003, 185(3);973-82
[PubMed:12533473]
[WorldCat.org]
[DOI]
(P p)
Szymon Krzywda, Andrzej M Brzozowski, Chandra Verma, Kiyonobu Karata, Teru Ogura, Anthony J Wilkinson
The crystal structure of the AAA domain of the ATP-dependent protease FtsH of Escherichia coli at 1.5 A resolution.
Structure: 2002, 10(8);1073-83
[PubMed:12176385]
[WorldCat.org]
[DOI]
(P p)
R S Prajapati, T Ogura, S M Cutting
Structural and functional studies on an FtsH inhibitor from Bacillus subtilis.
Biochim Biophys Acta: 2000, 1475(3);353-9
[PubMed:10913836]
[WorldCat.org]
[DOI]
(P p)
W Wehrl, M Niederweis, W Schumann
The FtsH protein accumulates at the septum of Bacillus subtilis during cell division and sporulation.
J Bacteriol: 2000, 182(13);3870-3
[PubMed:10851010]
[WorldCat.org]
[DOI]
(P p)
W Schumann
FtsH--a single-chain charonin?
FEMS Microbiol Rev: 1999, 23(1);1-11
[PubMed:10077851]
[WorldCat.org]
[DOI]
(P p)
M M Nakano, Y P Dailly, P Zuber, D P Clark
Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth.
J Bacteriol: 1997, 179(21);6749-55
[PubMed:9352926]
[WorldCat.org]
[DOI]
(P p)
Elena Lysenko, Teru Ogura, Simon M Cutting
Characterization of the ftsH gene of Bacillus subtilis.
Microbiology (Reading): 1997, 143 ( Pt 3);971-978
[PubMed:9084181]
[WorldCat.org]
[DOI]
(P p)
E Deuerling, A Mogk, C Richter, M Purucker, W Schumann
The ftsH gene of Bacillus subtilis is involved in major cellular processes such as sporulation, stress adaptation and secretion.
Mol Microbiol: 1997, 23(5);921-33
[PubMed:9076729]
[WorldCat.org]
[DOI]
(P p)
E Deuerling, B Paeslack, W Schumann
The ftsH gene of Bacillus subtilis is transiently induced after osmotic and temperature upshift.
J Bacteriol: 1995, 177(14);4105-12
[PubMed:7608085]
[WorldCat.org]
[DOI]
(P p)