Difference between revisions of "KtrA"
(→Extended information on the protein) |
|||
| Line 121: | Line 121: | ||
* '''[[Sigma factor]]:''' | * '''[[Sigma factor]]:''' | ||
| − | * '''Regulation:''' | + | * '''Regulation:''' |
| − | |||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| − | ** expression is controlled via termination antitermination by the [[ydaO riboswitch]] {{PubMed| | + | ** expression is controlled via termination antitermination by the [[ydaO riboswitch]], expression is switched off upon binding od c-di-AMP {{PubMed|24141192,20511502}} |
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 154: | Line 153: | ||
=References= | =References= | ||
| − | <pubmed>12562800,15096624,16321950, 20511502, 23086297 23598340 23671116 16990138 </pubmed> | + | <pubmed>12562800,15096624,16321950, 20511502, 23086297 23598340 23671116 16990138 24141192</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 17:08, 14 December 2013
- Description: high affinity potassium transporter KtrA-KtrB, peripheric membrane component (proton symport)
| Gene name | ktrA |
| Synonyms | yuaA |
| Essential | no |
| Product | high affinity potassium transporter KtrA-KtrB, peripheric membrane component (proton symport) |
| Function | potassium uptake |
| Gene expression levels in SubtiExpress: ktrA | |
| Interactions involving this protein in SubtInteract: KtrA | |
| Metabolic function and regulation of this protein in SubtiPathways: Metal ion homeostasis, Stress | |
| MW, pI | 24 kDa, 5.981 |
| Gene length, protein length | 666 bp, 222 aa |
| Immediate neighbours | bslA, ktrB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 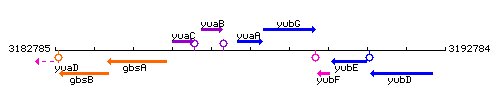
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed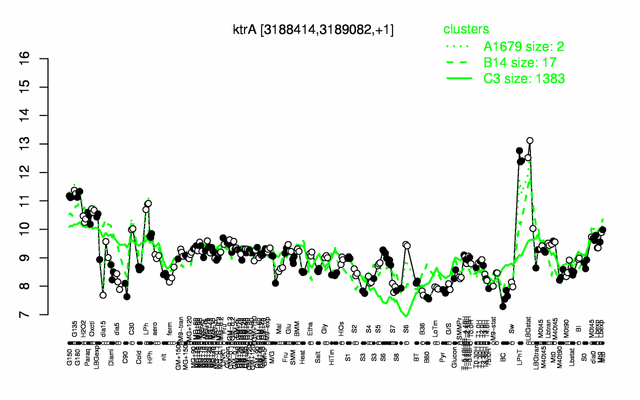
| |
Contents
Categories containing this gene/protein
transporters/ other, metal ion homeostasis (K, Na, Ca, Mg), coping with hyper-osmotic stress, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU31090
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family:
- Paralogous protein(s): KtrC
Extended information on the protein
- Kinetic information:
- Domains:
- contains a RCK_N domain at the N-terminus (according to UniProt, [2])
- contains a c-di-AMP-binding RCK_C domain at the C-terminus (according to UniProt, [3]) PubMed
- Modification:
- Effectors of protein activity:
- the protein binds c-di-AMP PubMed
- Localization: peripheral membrane protein PubMed
Database entries
- UniProt: O32080
- KEGG entry: [4]
- E.C. number:
Additional information
Expression and regulation
- Regulation:
- Regulatory mechanism:
- expression is controlled via termination antitermination by the ydaO riboswitch, expression is switched off upon binding od c-di-AMP PubMed
- Additional information:
Biological materials
- Mutant:
- 1A954 ( ktrA::kan), PubMed, available at BGSC
- GHB1 (D(ktrA-ktrB)::aphA3), available in Erhard Bremer's lab
- GP92 (D(ktrA-ktrB)::aphA3), available in Jörg Stülke's lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Erhard Bremer, University of Marburg, Germany homepage
Your additional remarks
References