Difference between revisions of "UvrA"
(→Original publications) |
|||
| Line 142: | Line 142: | ||
<pubmed>16464004 15927210 7801120 22933559 </pubmed> | <pubmed>16464004 15927210 7801120 22933559 </pubmed> | ||
== Original publications == | == Original publications == | ||
| − | <pubmed>11751826,,8226626, 24118570</pubmed> | + | <pubmed>11751826, 21145481,8226626, 24118570</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 16:35, 8 March 2014
- Description: excinuclease ABC (subunit A)
| Gene name | uvrA |
| Synonyms | |
| Essential | no |
| Product | excinuclease ABC (subunit A) |
| Function | DNA repair after UV damage |
| Gene expression levels in SubtiExpress: uvrA | |
| Interactions involving this protein in SubtInteract: UvrA | |
| MW, pI | 105 kDa, 5.843 |
| Gene length, protein length | 2871 bp, 957 aa |
| Immediate neighbours | yvzB, uvrB |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 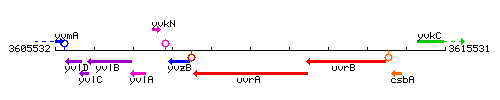
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU35160
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: UvrA subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: cytoplasm (according to Swiss-Prot)
Database entries
- Structure: 2R6F (Geobacillus stearothermophilus)
- UniProt: O34863
- KEGG entry: [3]
- E.C. number:
Additional information
Expression and regulation
- Additional information:
- during sporulation expressed both in the forespore and the mother cell PubMed
Biological materials
- mutant: GP1175 (del uvrAB::ermC) (available in the Stülke lab)
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications