Difference between revisions of "Mfd"
| Line 1: | Line 1: | ||
| − | * '''Description:''' transcription-repair coupling factor, eliminates genetic damage from transcriptionally active genes during [[sporulation]] <br/><br/> | + | * '''Description:''' [[transcription]]-repair coupling factor, eliminates genetic damage from transcriptionally active genes during [[sporulation]] <br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
| Line 10: | Line 10: | ||
|style="background:#ABCDEF;" align="center"| '''Essential''' || no | |style="background:#ABCDEF;" align="center"| '''Essential''' || no | ||
|- | |- | ||
| − | |style="background:#ABCDEF;" align="center"| '''Product''' || transcription-repair coupling factor | + | |style="background:#ABCDEF;" align="center"| '''Product''' || [[transcription]]-repair coupling factor |
|- | |- | ||
|style="background:#ABCDEF;" align="center"|'''Function''' || promotes strand-specific DNA repair by displacing | |style="background:#ABCDEF;" align="center"|'''Function''' || promotes strand-specific DNA repair by displacing | ||
| Line 61: | Line 61: | ||
* in an ''mfd'' knock-out, the cell's ability to accumulate adaptive mutations in stationary phase is depressed. [http://www.pubmed.com/16950921 PubMed] | * in an ''mfd'' knock-out, the cell's ability to accumulate adaptive mutations in stationary phase is depressed. [http://www.pubmed.com/16950921 PubMed] | ||
* increased UV-induced mutagenesis via [[PolY1]]/ [[PolY2]]-mediated translesion synthesis {{PubMed|24118570}} | * increased UV-induced mutagenesis via [[PolY1]]/ [[PolY2]]-mediated translesion synthesis {{PubMed|24118570}} | ||
| + | * the mutation suppresses the mucoid phenotype of ''[[motA]]'' or ''[[motB]]'' mutants {{PubMed|24296669}} | ||
=== Database entries === | === Database entries === | ||
| Line 86: | Line 87: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 131: | Line 132: | ||
=Biological materials = | =Biological materials = | ||
| − | + | * '''Mutant:''' GP1167 (del ermC), available in [[Jörg Stülke]]'s lab | |
| − | * '''Mutant:''' GP1167 (del ermC), available in [[Stülke]] lab | ||
* '''Expression vector:''' | * '''Expression vector:''' | ||
| Line 138: | Line 138: | ||
* '''lacZ fusion:''' | * '''lacZ fusion:''' | ||
| − | * '''GFP fusion:''' GP1510 (spc, based on [[pGP1870]], [[pGP1389]]-derivative ), available in | + | * '''GFP fusion:''' GP1510 (spc, based on [[pGP1870]], [[pGP1389]]-derivative ), available in [[Jörg Stülke]]'s lab |
| − | * '''YFP fusion:''' GP1511 (spc, based on [[pGP1871]], [[pGP1389]]-derivative ), available in | + | * '''YFP fusion:''' GP1511 (spc, based on [[pGP1871]], [[pGP1389]]-derivative ), available in [[Jörg Stülke]]'s lab |
* '''two-hybrid system:''' | * '''two-hybrid system:''' | ||
| Line 156: | Line 156: | ||
<pubmed>17239578 15063847 17917874 8811173 7968917 15720542 16464004 19023283 22201950 22749141 22933559 </pubmed> | <pubmed>17239578 15063847 17917874 8811173 7968917 15720542 16464004 19023283 22201950 22749141 22933559 </pubmed> | ||
== Original publications == | == Original publications == | ||
| − | <pubmed>8594198,20435731 ,104170,16950921,9535092, 16469698 21699902 22178973 22248542 22960746 24118570 21710567,20702425</pubmed> | + | <pubmed>8594198,20435731 ,104170,16950921,9535092, 16469698 21699902 22178973 22248542 22960746 24118570 21710567,20702425 24296669</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 19:59, 4 December 2013
- Description: transcription-repair coupling factor, eliminates genetic damage from transcriptionally active genes during sporulation
| Gene name | mfd |
| Synonyms | |
| Essential | no |
| Product | transcription-repair coupling factor |
| Function | promotes strand-specific DNA repair by displacing
RNA polymerase stalled at a nucleotide lesion and directing the (A)BC excinuclease to the RNA damage site |
| Gene expression levels in SubtiExpress: mfd
RNA polymerase stalled at a nucleotide lesion and directing the (A)BC excinuclease to the RNA damage site | |
| Interactions involving this protein in SubtInteract: Mfd | |
| MW, pI | 133 kDa, 5.367 |
| Gene length, protein length | 3531 bp, 1177 aa |
| Immediate neighbours | fin, spoVT |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 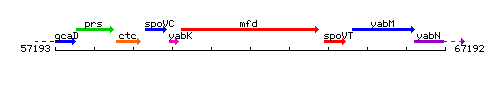
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
DNA repair/ recombination, transcription
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00550
Phenotypes of a mutant
- in an mfd knock-out, the cell's ability to accumulate adaptive mutations in stationary phase is depressed. PubMed
- increased UV-induced mutagenesis via PolY1/ PolY2-mediated translesion synthesis PubMed
- the mutation suppresses the mucoid phenotype of motA or motB mutants PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- promotes strand-specific DNA repair by displacing RNA polymerase stalled at a nucleotide lesion and directing the (A)BC excinuclease to the RNA damage site
- is required for roadblock transcription repression by transcription factors with binding sites downstream of the promoter (as for CcpA PubMed and CodY PubMed)
- Protein family:
- Paralogous protein(s): RecG
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
Database entries
- Structure:
- UniProt: P37474
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulation:
- Regulatory mechanism:
- Additional information:
- expressed throughout growth and sporulation, during sporulation expressed both in the forespore and the mother cell PubMed
Biological materials
- Mutant: GP1167 (del ermC), available in Jörg Stülke's lab
- Expression vector:
- lacZ fusion:
- GFP fusion: GP1510 (spc, based on pGP1870, pGP1389-derivative ), available in Jörg Stülke's lab
- YFP fusion: GP1511 (spc, based on pGP1871, pGP1389-derivative ), available in Jörg Stülke's lab
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Original publications