Difference between revisions of "Ctc"
| Line 18: | Line 18: | ||
|colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=Ctc Ctc] | |colspan="2" style="background:#FAF8CC;" align="center"| '''Interactions involving this protein in [http://subtiwiki.uni-goettingen.de/interact/ ''Subt''Interact]''': [http://subtiwiki.uni-goettingen.de/interact/index.php?protein=Ctc Ctc] | ||
|- | |- | ||
| − | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/ | + | |colspan="2" style="background:#FAF8CC;" align="center"| '''Metabolic function and regulation of this protein in [[SubtiPathways|''Subti''Pathways]]: <br/>[http://subtiwiki.uni-goettingen.de/subtipathways/search.php?enzyme=ctc ctc]''' |
|- | |- | ||
|style="background:#ABCDEF;" align="center"| '''MW, pI''' || 21 kDa, 4.216 | |style="background:#ABCDEF;" align="center"| '''MW, pI''' || 21 kDa, 4.216 | ||
Revision as of 09:58, 7 January 2014
- Description: general stress protein, similar to ribosomal protein L25
| Gene name | ctc |
| Synonyms | |
| Essential | no |
| Product | ribosomal protein |
| Function | unknown |
| Gene expression levels in SubtiExpress: ctc | |
| Interactions involving this protein in SubtInteract: Ctc | |
| Metabolic function and regulation of this protein in SubtiPathways: ctc | |
| MW, pI | 21 kDa, 4.216 |
| Gene length, protein length | 612 bp, 204 aa |
| Immediate neighbours | prs, spoVC |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 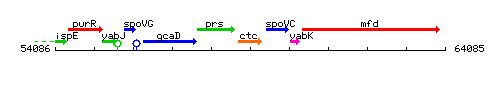
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed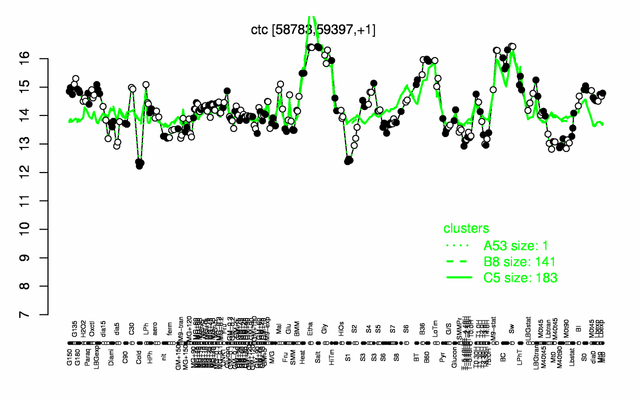
| |
Contents
Categories containing this gene/protein
translation, general stress proteins (controlled by SigB)
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU00520
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- Protein family: Ctc subfamily (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization: in the ribosome (large subunit) PubMed
Database entries
- UniProt: P14194
- KEGG entry: [3]
- E.C. number:
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulatory mechanism: alternative sigma factor, SigB PubMed
- Additional information: subject to Clp-dependent proteolysis upon glucose starvation PubMed
Biological materials
- Mutant: GP500(spc), available in Stülke lab
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody: available in Stülke lab
Labs working on this gene/protein
Your additional remarks
References
Reviews
Additional reviews: PubMed
Original publications
Genki Akanuma, Hideaki Nanamiya, Yousuke Natori, Koichi Yano, Shota Suzuki, Shuya Omata, Morio Ishizuka, Yasuhiko Sekine, Fujio Kawamura
Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation.
J Bacteriol: 2012, 194(22);6282-91
[PubMed:23002217]
[WorldCat.org]
[DOI]
(I p)
Dirk Höper, Uwe Völker, Michael Hecker
Comprehensive characterization of the contribution of individual SigB-dependent general stress genes to stress resistance of Bacillus subtilis.
J Bacteriol: 2005, 187(8);2810-26
[PubMed:15805528]
[WorldCat.org]
[DOI]
(P p)
Matthias Schmalisch, Ines Langbein, Jörg Stülke
The general stress protein Ctc of Bacillus subtilis is a ribosomal protein.
J Mol Microbiol Biotechnol: 2002, 4(5);495-501
[PubMed:12432960]
[WorldCat.org]
(P p)
R Fedorov, V Meshcheryakov, G Gongadze, N Fomenkova, N Nevskaya, M Selmer, M Laurberg, O Kristensen, S Al-Karadaghi, A Liljas, M Garber, S Nikonov
Structure of ribosomal protein TL5 complexed with RNA provides new insights into the CTC family of stress proteins.
Acta Crystallogr D Biol Crystallogr: 2001, 57(Pt 7);968-76
[PubMed:11418764]
[WorldCat.org]
[DOI]
(P p)
I Hilden, B N Krath, B Hove-Jensen
Tricistronic operon expression of the genes gcaD (tms), which encodes N-acetylglucosamine 1-phosphate uridyltransferase, prs, which encodes phosphoribosyl diphosphate synthetase, and ctc in vegetative cells of Bacillus subtilis.
J Bacteriol: 1995, 177(24);7280-4
[PubMed:8522540]
[WorldCat.org]
[DOI]
(P p)
C L Truitt, E A Weaver, W G Haldenwang
Effects on growth and sporulation of inactivation of a Bacillus subtilis gene (ctc) transcribed in vitro by minor vegetative cell RNA polymerases (E-sigma 37, E-sigma 32).
Mol Gen Genet: 1988, 212(1);166-71
[PubMed:2836704]
[WorldCat.org]
[DOI]
(P p)
C Ray, M Igo, W Shafer, R Losick, C P Moran
Suppression of ctc promoter mutations in Bacillus subtilis.
J Bacteriol: 1988, 170(2);900-7
[PubMed:3123466]
[WorldCat.org]
[DOI]
(P p)
M M Igo, R Losick
Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis.
J Mol Biol: 1986, 191(4);615-24
[PubMed:3100810]
[WorldCat.org]
[DOI]
(P p)
J F Ollington, W G Haldenwang, T V Huynh, R Losick
Developmentally regulated transcription in a cloned segment of the Bacillus subtilis chromosome.
J Bacteriol: 1981, 147(2);432-42
[PubMed:6790515]
[WorldCat.org]
[DOI]
(P p)
W G Haldenwang, R Losick
A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis.
Nature: 1979, 282(5736);256-60
[PubMed:116131]
[WorldCat.org]
[DOI]
(P p)