Difference between revisions of "GlmS"
(→Other Original Publications) |
|||
| Line 79: | Line 79: | ||
* '''Kinetic information:''' | * '''Kinetic information:''' | ||
| − | * '''Domains:''' | + | * '''[[Domains]]:''' |
* '''Modification:''' | * '''Modification:''' | ||
| − | * ''' | + | * '''[[Cofactors]]:''' |
* '''Effectors of protein activity:''' | * '''Effectors of protein activity:''' | ||
| Line 149: | Line 149: | ||
==The ''glmS'' Ribozyme== | ==The ''glmS'' Ribozyme== | ||
| − | <pubmed>18079181 ,16484375, 16784238 ,15096624 , 16990543 ,17114942 ,16484375 , 15029187, 17283212 , 16298301, 19228039 21317896 21395279 </pubmed> | + | <pubmed>18079181 ,16484375, 16784238 ,15096624 , 16990543 ,17114942 ,16484375 , 15029187, 17283212 , 16298301, 19228039 21317896 21395279 24318896 </pubmed> |
==Other Original Publications== | ==Other Original Publications== | ||
<pubmed> 14343123 17981983 ,11160890, 18295797 20525796 23192352 23876412 22211522</pubmed> | <pubmed> 14343123 17981983 ,11160890, 18295797 20525796 23192352 23876412 22211522</pubmed> | ||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 18:11, 13 December 2013
- Description: glutamine-fructose-6-phosphate transaminase
| Gene name | glmS |
| Synonyms | gcaA, ybxD |
| Essential | yes PubMed |
| Product | glutamine-fructose-6-phosphate transaminase |
| Function | cell wall synthesis |
| Gene expression levels in SubtiExpress: glmS | |
| Metabolic function and regulation of this protein in SubtiPathways: Murein recycling | |
| MW, pI | 65 kDa, 4.796 |
| Gene length, protein length | 1800 bp, 600 aa |
| Immediate neighbours | glmM, alkA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 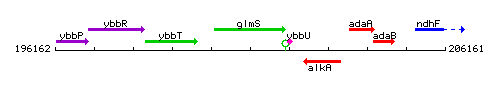
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
cell wall synthesis, biosynthesis of cell wall components, essential genes
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU01780
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: L-glutamine + D-fructose 6-phosphate = L-glutamate + D-glucosamine 6-phosphate (according to Swiss-Prot)
- Protein family:
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Modification:
- Effectors of protein activity:
- Localization:
- cytoplasm (according to Swiss-Prot)
Database entries
- UniProt: P39754
- KEGG entry: [2]
- E.C. number: 2.6.1.16
Additional information
- subject to Clp-dependent proteolysis upon glucose starvation PubMed
Expression and regulation
- Regulation:
- Regulatory mechanism: glmS ribozyme: glucosamine 6-phosphate binds the leader mRNA, and a riboswitch with ribozyme activity cleaves off the glmS section from the mRNA, resulting in stopp of transcript elongation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Wade Winkler, University of Texas, USA, Homepage
Your additional remarks
References
Reviews
The glmS Ribozyme
Other Original Publications