Difference between revisions of "CggR"
| Line 114: | Line 114: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=cggR_3482752_3483774_-1 cggR] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=cggR_3482752_3483774_-1 cggR] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' [[SigA]] {{PubMed|11489127}} | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|11489127}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| Line 133: | Line 133: | ||
* '''Mutant:''' | * '''Mutant:''' | ||
| − | ** GP311 (in frame deletion), available in [[Stülke]] lab | + | ** GP311 (in frame deletion), available in [[Jörg Stülke]]'s lab |
** SM-NB7 (cggR-spc), available in [[Anne Galinier]]'s and [[Boris Görke]]'s labs | ** SM-NB7 (cggR-spc), available in [[Anne Galinier]]'s and [[Boris Görke]]'s labs | ||
| − | * '''Expression vector:''' pGP705 (N-terminal His-tag, in [[pWH844]]), available in [[Stülke]] lab | + | * '''Expression vector:''' pGP705 (N-terminal His-tag, in [[pWH844]]), available in [[Jörg Stülke]]'s lab |
| − | * '''lacZ fusion:''' pGP504 (in [[pAC7]]), pGP509 (in [[pAC6]]), available in [[Stülke]] lab | + | * '''lacZ fusion:''' pGP504 (in [[pAC7]]), pGP509 (in [[pAC6]]), available in [[Jörg Stülke]]'s lab |
* '''GFP fusion:''' | * '''GFP fusion:''' | ||
| − | * '''Antibody:''' available in [[Stülke]] lab | + | * '''Antibody:''' available in [[Jörg Stülke]]'s lab |
=Labs working on this gene/protein= | =Labs working on this gene/protein= | ||
| Line 155: | Line 155: | ||
==Original Publications== | ==Original Publications== | ||
| − | + | ||
| − | <pubmed> 22190493 23280112 | + | <pubmed>20462860 22190493 23280112 21815947 19193632,12850135, 12622823,18186488,10799476,11489127, 12123463,12634343, 18052209 , 17293407 20361740 20444087 18554327,</pubmed> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 15:25, 13 July 2013
- Description: repressor of the glycolytic gapA operon, DeoR family
| Gene name | cggR |
| Synonyms | yvbQ |
| Essential | no |
| Product | central glycolytic genes regulator |
| Function | transcriptional regulator |
| Gene expression levels in SubtiExpress: cggR | |
| Metabolic function and regulation of this protein in SubtiPathways: Central C-metabolism | |
| MW, pI | 37,2 kDa,5.68 |
| Gene length, protein length | 1020 bp, 340 amino acids |
| Immediate neighbours | gapA, araE |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 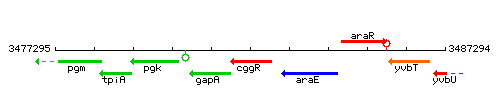
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
carbon core metabolism, transcription factors and their control, regulators of core metabolism
This gene is a member of the following regulons
The CggR regulon:
The gene
Basic information
- Locus tag: BSU33950
Phenotypes of a mutant
Database entries
- DBTBS entry: [1]
- SubtiList entry: [2]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: transcription repression of the glycolytic gapA operon
- Protein family: sorC transcriptional regulatory family (according to Swiss-Prot)
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- DNA binding domain (H-T-H motif) (37–56)
- Modification:
- Cofactor(s):
- Effectors of protein activity: fructose 1.6-bisphosphate PubMed and dihydroxyacetone phosphate, glucose-6-phosphate and fructose-6-phosphate PubMed act as inducer and result in release of CggR from the DNA
- Interactions:
- active as dimer (according to PubMed)
- Localization: cytoplasma PubMed
Database entries
- Structure: 2OKG ( effector binding domain), 3BXH (in complex with fructose-6-phosphate), complex with Fructose-6-Phosphate NCBI, effector binding domain NCBI
- UniProt: O32253
- KEGG entry: [3]
Additional information
Expression and regulation
- Database entries: DBTBS
- Additional information:
- The primary mRNAs of the operon are highly unstable. The primary mRNA is subject to processing at the very end of the cggR open reading frame. This results in stable mature gapA and gapA-pgk-tpiA-pgm-eno mRNAs. PubMed The processing event requires the RNase Y PubMed.
- The intracellular concentration of CggR is about 230 nM (according to PubMed).
- The accumulation of the cggR-gapA mRNA is strongly dependent on the presence of the YkzW peptide, due to stabilization of the mRNA PubMed.
- the mRNA is substantially stabilized upon depletion of RNase Y PubMed
Biological materials
- Mutant:
- GP311 (in frame deletion), available in Jörg Stülke's lab
- SM-NB7 (cggR-spc), available in Anne Galinier's and Boris Görke's labs
- Expression vector: pGP705 (N-terminal His-tag, in pWH844), available in Jörg Stülke's lab
- lacZ fusion: pGP504 (in pAC7), pGP509 (in pAC6), available in Jörg Stülke's lab
- GFP fusion:
- Antibody: available in Jörg Stülke's lab
Labs working on this gene/protein
Stephane Aymerich, Microbiology and Molecular Genetics, INRA Paris-Grignon, France
Your additional remarks
References
Reviews
Original Publications