Difference between revisions of "Cca"
| Line 35: | Line 35: | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
<br/><br/><br/><br/> | <br/><br/><br/><br/> | ||
| − | |||
| − | |||
| − | |||
| − | |||
<br/><br/> | <br/><br/> | ||
| Line 46: | Line 42: | ||
= This gene is a member of the following [[regulons]] = | = This gene is a member of the following [[regulons]] = | ||
| + | {{SubtiWiki regulon|[[Spx regulon]]}} | ||
| Line 113: | Line 110: | ||
* '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=cca_2355880_2357073_-1 cca] {{PubMed|22383849}} | * '''Expression browser:''' [http://genome.jouy.inra.fr/cgi-bin/seb/viewdetail.py?id=cca_2355880_2357073_-1 cca] {{PubMed|22383849}} | ||
| − | * '''Sigma factor:''' | + | * '''[[Sigma factor]]:''' [[SigA]] {{PubMed|23894131}} |
* '''Regulation:''' | * '''Regulation:''' | ||
| + | ** induced by diamide stess (thiol depletion) ([[Spx]]) {{PubMed|23894131}} | ||
* '''Regulatory mechanism:''' | * '''Regulatory mechanism:''' | ||
| + | ** [[Spx]]: transcription activation {{PubMed|23894131}} | ||
* '''Additional information:''' | * '''Additional information:''' | ||
| Line 145: | Line 144: | ||
<pubmed>17179213,12526808,20360175 ,9829937, 16109934 </pubmed> | <pubmed>17179213,12526808,20360175 ,9829937, 16109934 </pubmed> | ||
==Operon and expression== | ==Operon and expression== | ||
| − | <pubmed> 20308541 </pubmed> | + | <pubmed> 20308541 23894131</pubmed> |
[[Category:Protein-coding genes]] | [[Category:Protein-coding genes]] | ||
Revision as of 12:14, 1 August 2013
- Description: tRNA nucleotidyltransferase, maturation of the single-copy tRNACys, which lacks an encoded CCA 3' end
| Gene name | cca |
| Synonyms | papS, ypjI |
| Essential | yes PubMed |
| Product | tRNA nucleotidyltransferase |
| Function | tRNA modification |
| Gene expression levels in SubtiExpress: cca | |
| MW, pI | 45 kDa, 8.041 |
| Gene length, protein length | 1191 bp, 397 aa |
| Immediate neighbours | birA, bshA |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 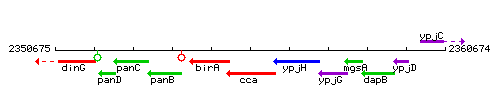
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed
| |
Contents
Categories containing this gene/protein
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU22450
Phenotypes of a mutant
essential PubMed
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity: ATP + tRNA(n) = diphosphate + tRNA(n+1) (according to Swiss-Prot)
- Protein family: the protein is similar to the E. coli poly(A) polymerase
- Paralogous protein(s):
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
Database entries
- Structure: 1MIV (Geobacillus stearothermophilus 44% identity)
- UniProt: P42977
- KEGG entry: [2]
- E.C. number: 2.7.7.25
Additional information
Expression and regulation
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Your additional remarks
References
Reviews
Heike Betat, Christiane Rammelt, Mario Mörl
tRNA nucleotidyltransferases: ancient catalysts with an unusual mechanism of polymerization.
Cell Mol Life Sci: 2010, 67(9);1447-63
[PubMed:20155482]
[WorldCat.org]
[DOI]
(I p)
Stefan Vörtler, Mario Mörl
tRNA-nucleotidyltransferases: highly unusual RNA polymerases with vital functions.
FEBS Lett: 2010, 584(2);297-302
[PubMed:19883645]
[WorldCat.org]
[DOI]
(I p)
Anne Neuenfeldt, Andrea Just, Heike Betat, Mario Mörl
Evolution of tRNA nucleotidyltransferases: a small deletion generated CC-adding enzymes.
Proc Natl Acad Sci U S A: 2008, 105(23);7953-8
[PubMed:18523015]
[WorldCat.org]
[DOI]
(I p)
Yong Xiong, Thomas A Steitz
A story with a good ending: tRNA 3'-end maturation by CCA-adding enzymes.
Curr Opin Struct Biol: 2006, 16(1);12-7
[PubMed:16364630]
[WorldCat.org]
[DOI]
(P p)
Original publications
Operon and expression