Difference between revisions of "MreBH"
| Line 1: | Line 1: | ||
| − | * '''Description:''' [[cell shape]]-determining protein, forms filaments, the polymers control/restrict the mobility of the cell wall elongation enzyme | + | * '''Description:''' [[cell shape]]-determining protein, forms filaments, the polymers control/restrict the mobility of the cell wall elongation enzyme complex<br/><br/> |
{| align="right" border="1" cellpadding="2" | {| align="right" border="1" cellpadding="2" | ||
Revision as of 08:47, 7 June 2013
- Description: cell shape-determining protein, forms filaments, the polymers control/restrict the mobility of the cell wall elongation enzyme complex
| Gene name | mreBH |
| Synonyms | |
| Essential | no |
| Product | cell shape-determining protein |
| Function | cell shape determation |
| Gene expression levels in SubtiExpress: mreBH | |
| Interactions involving this protein in SubtInteract: MreBH | |
| MW, pI | 35 kDa, 5.239 |
| Gene length, protein length | 1005 bp, 335 aa |
| Immediate neighbours | ykpC, abh |
| Sequences | Protein DNA DNA_with_flanks |
Genetic context 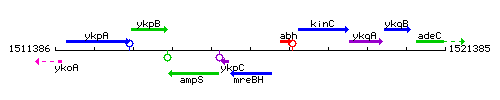
This image was kindly provided by SubtiList
| |
Expression at a glance PubMed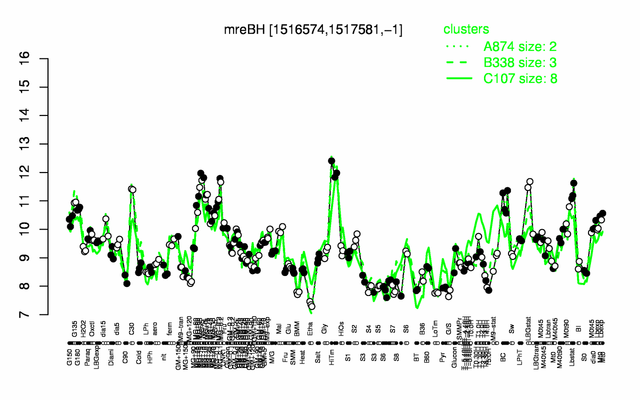
| |
Contents
Categories containing this gene/protein
cell shape, heat shock proteins, membrane proteins
This gene is a member of the following regulons
The gene
Basic information
- Locus tag: BSU14470
Phenotypes of a mutant
Database entries
- DBTBS entry: no entry
- SubtiList entry: [1]
Additional information
The protein
Basic information/ Evolution
- Catalyzed reaction/ biological activity:
- forms ring-shaped filaments in a heterologous system PubMed
- Protein family: ftsA/mreB family (according to Swiss-Prot)
Extended information on the protein
- Kinetic information:
- Domains:
- Modification:
- Cofactor(s):
- Effectors of protein activity:
- Localization:
- during logarithmic growth, MreBH forms discrete patches thst move processively along peripheral tracks perpendicular to the cell axis PubMed
- forms transverse bands as cells enter the stationary phase PubMed
- close to the inner surface of the cytoplasmic membrane PubMed
- reports on helical structures formed by MreBH PubMed seem to be misinterpretation of data PubMed
- normal localization depends on the presence of glucolipids, MreB forms irregular clusters in an ugtP mutant PubMed
Database entries
- Structure:
- UniProt: P39763
- KEGG entry: [2]
- E.C. number:
Additional information
Expression and regulation
- Operon:
- Regulatory mechanism:
- Additional information:
Biological materials
- Mutant:
- Expression vector:
- lacZ fusion:
- GFP fusion:
- two-hybrid system:
- Antibody:
Labs working on this gene/protein
Peter Graumann, Freiburg University, Germany homepage
Your additional remarks
References
Localization
Other original publications
Felix Dempwolff, Christian Reimold, Michael Reth, Peter L Graumann
Bacillus subtilis MreB orthologs self-organize into filamentous structures underneath the cell membrane in a heterologous cell system.
PLoS One: 2011, 6(11);e27035
[PubMed:22069484]
[WorldCat.org]
[DOI]
(I p)
Hervé Joël Defeu Soufo, Peter L Graumann
Bacillus subtilis MreB paralogues have different filament architectures and lead to shape remodelling of a heterologous cell system.
Mol Microbiol: 2010, 78(5);1145-58
[PubMed:21091501]
[WorldCat.org]
[DOI]
(I p)
Yoshikazu Kawai, Kei Asai, Jeffery Errington
Partial functional redundancy of MreB isoforms, MreB, Mbl and MreBH, in cell morphogenesis of Bacillus subtilis.
Mol Microbiol: 2009, 73(4);719-31
[PubMed:19659933]
[WorldCat.org]
[DOI]
(I p)
Kathrin Schirner, Jeff Errington
Influence of heterologous MreB proteins on cell morphology of Bacillus subtilis.
Microbiology (Reading): 2009, 155(Pt 11);3611-3621
[PubMed:19643765]
[WorldCat.org]
[DOI]
(I p)
Chi-Ling Tseng, Gwo-Chyuan Shaw
Genetic evidence for the actin homolog gene mreBH and the bacitracin resistance gene bcrC as targets of the alternative sigma factor SigI of Bacillus subtilis.
J Bacteriol: 2008, 190(5);1561-7
[PubMed:18156261]
[WorldCat.org]
[DOI]
(I p)
Rut Carballido-López, Alex Formstone, Ying Li, S Dusko Ehrlich, Philippe Noirot, Jeff Errington
Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE.
Dev Cell: 2006, 11(3);399-409
[PubMed:16950129]
[WorldCat.org]
[DOI]
(P p)